Above the Epitranscriptome: RNA Modifications and Stem Cell Identity
- PMID: 29958477
- PMCID: PMC6070936
- DOI: 10.3390/genes9070329
Above the Epitranscriptome: RNA Modifications and Stem Cell Identity
Abstract
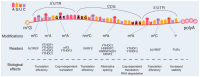
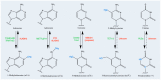
Sequence databases and transcriptome-wide mapping have revealed different reversible and dynamic chemical modifications of the nitrogen bases of RNA molecules. Modifications occur in coding RNAs and noncoding-RNAs post-transcriptionally and they can influence the RNA structure, metabolism, and function. The result is the expansion of the variety of the transcriptome. In fact, depending on the type of modification, RNA molecules enter into a specific program exerting the role of the player or/and the target in biological and pathological processes. Many research groups are exploring the role of RNA modifications (alias epitranscriptome) in cell proliferation, survival, and in more specialized activities. More recently, the role of RNA modifications has been also explored in stem cell biology. Our understanding in this context is still in its infancy. Available evidence addresses the role of RNA modifications in self-renewal, commitment, and differentiation processes of stem cells. In this review, we will focus on five epitranscriptomic marks: N6-methyladenosine, N1-methyladenosine, 5-methylcytosine, Pseudouridine (Ψ) and Adenosine-to-Inosine editing. We will provide insights into the function and the distribution of these chemical modifications in coding RNAs and noncoding-RNAs. Mainly, we will emphasize the role of epitranscriptomic mechanisms in the biology of naïve, primed, embryonic, adult, and cancer stem cells.
Keywords: 5-methylcytosine; N1-methyladenosine; N6-methyladenosine; bioinformatics predictive tools; cancer stem cells; epigenetics; erasers proteins; mitochondrial ribosomal RNA; mitochondrial transfer RNA; naïve and primed stem cells; readers; stem cells self-renewal and differentiation; writers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Martino S., Morena F., Barola C., Bicchi I., Emiliani C. Proteomics and epigenetic mechanisms in stem cells. Curr. Proteom. 2014;11:193–209. doi: 10.2174/157016461103140922164050. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

