Significance Testing for Allelic Heterogeneity
- PMID: 29959179
- PMCID: PMC6116971
- DOI: 10.1534/genetics.118.301111
Significance Testing for Allelic Heterogeneity
Abstract
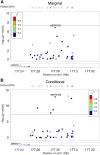
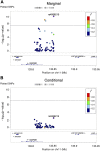
It is useful to detect allelic heterogeneity (AH), i.e., the presence of multiple causal SNPs in a locus, which, for example, may guide the development of new methods for fine mapping and determine how to interpret an appearing epistasis. In contrast to Mendelian traits, the existence and extent of AH for complex traits had been largely unknown until Hormozdiari et al. proposed a Bayesian method, called causal variants identification in associated regions (CAVIAR), and uncovered widespread AH in complex traits. However, there are several limitations with CAVIAR. First, it assumes a maximum number of causal SNPs in a locus, typically up to six, to save computing time; this assumption, as will be shown, may influence the outcome. Second, its computational time can be too demanding to be feasible since it examines all possible combinations of causal SNPs (under the assumed upper bound). Finally, it outputs a posterior probability of AH, which may be difficult to calibrate with a commonly used nominal significance level. Here, we introduce an intersection-union test (IUT) based on a joint/conditional regression model with all the SNPs in a locus to infer AH. We also propose two sequential IUT-based testing procedures to estimate the number of causal SNPs. Our proposed methods are applicable to not only individual-level genotypic and phenotypic data, but also genome-wide association study (GWAS) summary statistics. We provide numerical examples based on both simulated and real data, including large-scale schizophrenia (SCZ) and high-density lipoprotein (HDL) GWAS summary data sets, to demonstrate the effectiveness of the new methods. In particular, for both the SCZ and HDL data, our proposed IUT not only was faster, but also detected more AH loci than CAVIAR. Our proposed methods are expected to be useful in further uncovering the extent of AH in complex traits.
Keywords: CAVIAR; GWAS; conditional/joint modeling; intersection-union test; statistical power.
Copyright © 2018 by the Genetics Society of America.
Figures


References
-
- Berger, R. L., 1997 Likelihood ratio tests and intersection-union tests, pp. 225–237 in Advances in Statistical Decision Theory and Applications, edited by S. Panchapakesan and N. Balakrishnan. Birkhäuser, Boston.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

