Toward a Kinetic Understanding of Eukaryotic Translation
- PMID: 29959192
- PMCID: PMC6360857
- DOI: 10.1101/cshperspect.a032706
Toward a Kinetic Understanding of Eukaryotic Translation
Abstract
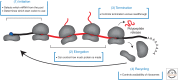
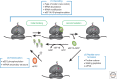
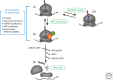
The eukaryotic translation pathway has been studied for more than four decades, but the molecular mechanisms that regulate each stage of the pathway are not completely defined. This is in part because we have very little understanding of the kinetic framework for the assembly and disassembly of pathway intermediates. Steps of the pathway are thought to occur in the subsecond to second time frame, but most assays to monitor these events require minutes to hours to complete. Understanding translational control in sufficient detail will therefore require the development of assays that can precisely monitor the kinetics of the translation pathway in real time. Here, we describe the translation pathway from the perspective of its kinetic parameters, discuss advances that are helping us move toward the goal of a rigorous kinetic understanding, and highlight some of the challenges that remain.
Copyright © 2019 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures

References
-
- Algire MA, Maag D, Lorsch JR. 2005. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell 20: 251–262. - PubMed
-
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. 2006. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 125: 1125–1136. - PubMed
-
- Aylett CH, Boehringer D, Erzberger JP, Schaefer T, Ban N. 2015. Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nat Struct Mol Biol 22: 269–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
