Environmental Controls on Soil Microbial Communities in a Seasonally Dry Tropical Forest
- PMID: 29959251
- PMCID: PMC6102984
- DOI: 10.1128/AEM.00342-18
Environmental Controls on Soil Microbial Communities in a Seasonally Dry Tropical Forest
Abstract
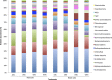
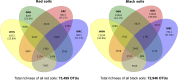
Several studies have shown that rainfall seasonality, soil heterogeneity, and increased nitrogen (N) deposition may have important effects on tropical forest function. However, the effects of these environmental controls on soil microbial communities in seasonally dry tropical forests are poorly understood. In a seasonally dry tropical forest in the Yucatan Peninsula (Mexico), we investigated the influence of soil heterogeneity (which results in two different soil types, black and red soils), rainfall seasonality (in two successive seasons, wet and dry), and 3 years of repeated N enrichment on soil chemical and microbiological properties, including bacterial gene content and community structure. The soil properties varied with the soil type and the sampling season but did not respond to N enrichment. Greater organic matter content in the black soils was associated with higher microbial biomass, enzyme activities, and abundances of genes related to nitrification (amoA) and denitrification (nirK and nirS) than were observed in the red soils. Rainfall seasonality was also associated with changes in soil microbial biomass and activity levels and N gene abundances. Actinobacteria, Proteobacteria, Firmicutes, and Acidobacteria were the most abundant phyla. Differences in bacterial community composition were associated with soil type and season and were primarily detected at higher taxonomic resolution, where specific taxa drive the separation of communities between soils. We observed that soil heterogeneity and rainfall seasonality were the main correlates of soil bacterial community structure and function in this tropical forest, likely acting through their effects on soil attributes, especially those related to soil organic matter and moisture content.IMPORTANCE Understanding the response of soil microbial communities to environmental factors is important for predicting the contribution of forest ecosystems to global environmental change. Seasonally dry tropical forests are characterized by receiving less than 1,800 mm of rain per year in alternating wet and dry seasons and by high heterogeneity in plant diversity and soil chemistry. For these reasons, N deposition may affect their soils differently than those in humid tropical forests. This study documents the influence of rainfall seasonality, soil heterogeneity, and N deposition on soil chemical and microbiological properties in a seasonally dry tropical forest. Our findings suggest that soil heterogeneity and rainfall seasonality are likely the main factors controlling soil bacterial community structure and function in this tropical forest. Nitrogen enrichment was likely too low to induce significant short-term effects on soil properties, because this tropical forest is not N limited.
Keywords: N deposition; functional N genes; nitrogen deposition; rainfall seasonality; soil bacterial community structure; soil heterogeneity; tropical dry forest.
Copyright © 2018 American Society for Microbiology.
Figures




Similar articles
-
[Seasonal changes of ammonia-oxidizing bacterial communities during tropical forest restoration].Ying Yong Sheng Tai Xue Bao. 2024 May;35(5):1242-1250. doi: 10.13287/j.1001-9332.202405.015. Ying Yong Sheng Tai Xue Bao. 2024. PMID: 38886422 Chinese.
-
Rainfall and labile carbon availability control litter nitrogen dynamics in a tropical dry forest.Oecologia. 2007 Jan;150(4):602-10. doi: 10.1007/s00442-006-0564-3. Epub 2006 Sep 22. Oecologia. 2007. PMID: 17024378
-
Distinct bacterial communities across a gradient of vegetation from a preserved Brazilian Cerrado.Antonie Van Leeuwenhoek. 2017 Apr;110(4):457-469. doi: 10.1007/s10482-016-0815-1. Epub 2017 Jan 6. Antonie Van Leeuwenhoek. 2017. PMID: 28062969
-
Forest Soil Bacteria: Diversity, Involvement in Ecosystem Processes, and Response to Global Change.Microbiol Mol Biol Rev. 2017 Apr 12;81(2):e00063-16. doi: 10.1128/MMBR.00063-16. Print 2017 Jun. Microbiol Mol Biol Rev. 2017. PMID: 28404790 Free PMC article. Review.
-
Ecology of Nitrogen Fixing, Nitrifying, and Denitrifying Microorganisms in Tropical Forest Soils.Front Microbiol. 2016 Jul 5;7:1045. doi: 10.3389/fmicb.2016.01045. eCollection 2016. Front Microbiol. 2016. PMID: 27468277 Free PMC article. Review.
Cited by
-
The response of root-zone soil bacterial community, metabolites, and soil properties of Sanyeqing medicinal plant varieties to anthracnose disease in reclaimed land, China.Heliyon. 2024 Aug 20;10(16):e36602. doi: 10.1016/j.heliyon.2024.e36602. eCollection 2024 Aug 30. Heliyon. 2024. PMID: 39258202 Free PMC article.
-
Diversity and Interactomics of Bacterial Communities Associated with Dominant Trees During Tropical Forest Recovery.Curr Microbiol. 2021 Sep;78(9):3417-3429. doi: 10.1007/s00284-021-02603-9. Epub 2021 Jul 10. Curr Microbiol. 2021. PMID: 34244846
-
Diversity of bacterial communities in wetlands of Calakmul Biosphere Reserve: a comparative analysis between conserved and semi-urbanized zones in pre-Mayan Train era.BMC Microbiol. 2024 Sep 28;24(1):376. doi: 10.1186/s12866-024-03523-x. BMC Microbiol. 2024. PMID: 39342129 Free PMC article.
-
The core bacteriobiome of Côte d'Ivoire soils across three vegetation zones.Front Microbiol. 2023 Aug 24;14:1220655. doi: 10.3389/fmicb.2023.1220655. eCollection 2023. Front Microbiol. 2023. PMID: 37692382 Free PMC article. Review.
-
The nesting preference of an invasive ant is associated with the cues produced by actinobacteria in soil.PLoS Pathog. 2020 Sep 10;16(9):e1008800. doi: 10.1371/journal.ppat.1008800. eCollection 2020 Sep. PLoS Pathog. 2020. PMID: 32913361 Free PMC article.
References
-
- Pajares S, Escalante AE, Noguez AM, García-Oliva F, Martínez-Piedragil C, Cram SS, Eguiarte LE, Souza V. 2016. Spatial heterogeneity of physicochemical properties explains differences in microbial composition in arid soils from Cuatro Cienegas, Mexico. PeerJ PrePrints 4:e2459. doi:10.7717/peerj.2459. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

