Pervasive genetic interactions modulate neurodevelopmental defects of the autism-associated 16p11.2 deletion in Drosophila melanogaster
- PMID: 29959322
- PMCID: PMC6026208
- DOI: 10.1038/s41467-018-04882-6
Pervasive genetic interactions modulate neurodevelopmental defects of the autism-associated 16p11.2 deletion in Drosophila melanogaster
Abstract
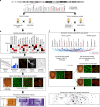
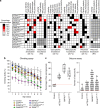
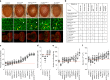
As opposed to syndromic CNVs caused by single genes, extensive phenotypic heterogeneity in variably-expressive CNVs complicates disease gene discovery and functional evaluation. Here, we propose a complex interaction model for pathogenicity of the autism-associated 16p11.2 deletion, where CNV genes interact with each other in conserved pathways to modulate expression of the phenotype. Using multiple quantitative methods in Drosophila RNAi lines, we identify a range of neurodevelopmental phenotypes for knockdown of individual 16p11.2 homologs in different tissues. We test 565 pairwise knockdowns in the developing eye, and identify 24 interactions between pairs of 16p11.2 homologs and 46 interactions between 16p11.2 homologs and neurodevelopmental genes that suppress or enhance cell proliferation phenotypes compared to one-hit knockdowns. These interactions within cell proliferation pathways are also enriched in a human brain-specific network, providing translational relevance in humans. Our study indicates a role for pervasive genetic interactions within CNVs towards cellular and developmental phenotypes.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM121907/GM/NIGMS NIH HHS/United States
- T32-GM102057/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)/International
- 22535/National Alliance for Research on Schizophrenia and Depression (NARSAD)/International
- 5-FY14-66/March of Dimes Foundation (March of Dimes)/International
- R01-GM121907/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)/International
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

