MicroRNA-9 inhibits growth and invasion of head and neck cancer cells and is a predictive biomarker of response to plerixafor, an inhibitor of its target CXCR4
- PMID: 29959873
- PMCID: PMC6275261
- DOI: 10.1002/1878-0261.12352
MicroRNA-9 inhibits growth and invasion of head and neck cancer cells and is a predictive biomarker of response to plerixafor, an inhibitor of its target CXCR4
Abstract
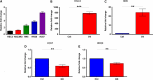
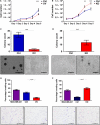
Head and neck squamous cell carcinomas (HNSCC) are associated with poor morbidity and mortality. Current treatment strategies are highly toxic and do not benefit over 50% of patients. There is therefore a crucial need for predictive and/or prognostic biomarkers to allow treatment stratification for individual patients. One class of biomarkers that has recently gained importance are microRNA (miRNA). MiRNA are small, noncoding molecules which regulate gene expression post-transcriptionally. We performed miRNA expression profiling of a cohort of head and neck tumours with known clinical outcomes. The results showed miR-9 to be significantly downregulated in patients with poor treatment outcome, indicating its role as a potential biomarker in HNSCC. Overexpression of miR-9 in HNSCC cell lines significantly decreased cellular proliferation and inhibited colony formation in soft agar. Conversely, miR-9 knockdown significantly increased both these features. Importantly, endogenous CXCR4 expression levels, a known target of miR-9, inversely correlated with miR-9 expression in a panel of HNSCC cell lines tested. Induced overexpression of CXCR4 in low expressing cells increased proliferation, colony formation and cell cycle progression. Moreover, CXCR4-specific ligand, CXCL12, enhanced cellular proliferation, migration, colony formation and invasion in CXCR4-overexpressing and similarly in miR-9 knockdown cells. CXCR4-specific inhibitor plerixafor abrogated the oncogenic phenotype of CXCR4 overexpression as well as miR-9 knockdown. Our data demonstrate a clear role for miR-9 as a tumour suppressor microRNA in HNSCC, and its role seems to be mediated through CXCR4 suppression. MiR-9 knockdown, similar to CXCR4 overexpression, significantly promoted aggressive HNSCC tumour cell characteristics. Our results suggest CXCR4-specific inhibitor plerixafor as a potential therapeutic agent, and miR-9 as a possible predictive biomarker of treatment response in HNSCC.
Keywords: CXCL12; CXCR4; MiR-9; head and neck cancer; plerixafor; tumour invasion.
© 2018 The Authors. Published by FEBS Press and John Wiley & Sons Ltd.
Figures





References
-
- Albert S, Riveiro ME, Halimi C, Hourseau M, Couvelard A, Serova M, Barry B, Raymond E and Faivre S (2013) Focus on the role of the CXCL12/CXCR4 chemokine axis in head and neck squamous cell carcinoma. Head Neck 35, 1819–1828. - PubMed
-
- Anonymous (2007) Plerixafor: AMD 3100, AMD3100, JM 3100, SDZ SID 791. Drugs R D 8, 113–119. - PubMed
-
- Balkwill F (2004) Cancer and the chemokine network. Nat Rev Cancer 4, 540–550. - PubMed
-
- Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman‐Gomez J, Prosper F and Garcia‐Foncillas J (2009) Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer 125, 2737–2743. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

