Human histone demethylase KDM6B can catalyse sequential oxidations
- PMID: 29961803
- PMCID: PMC6044289
- DOI: 10.1039/c8cc04057e
Human histone demethylase KDM6B can catalyse sequential oxidations
Abstract
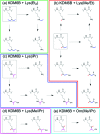
Jumonji domain-containing demethylases (JmjC-KDMs) catalyse demethylation of Nε-methylated lysines on histones and play important roles in gene regulation. We report selectivity studies on KDM6B (JMJD3), a disease-relevant JmjC-KDM, using synthetic lysine analogues. The results unexpectedly reveal that KDM6B accepts multiple Nε-alkylated lysine analogues, forming alcohol, aldehyde and carboxylic acid products.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

