Big Data Analysis of Genes Associated With Neuropsychiatric Disorders in an Alzheimer's Disease Animal Model
- PMID: 29962931
- PMCID: PMC6013555
- DOI: 10.3389/fnins.2018.00407
Big Data Analysis of Genes Associated With Neuropsychiatric Disorders in an Alzheimer's Disease Animal Model
Abstract
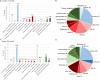
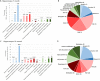
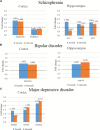
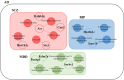
Alzheimer's disease is a neurodegenerative disease characterized by the impairment of cognitive function and loss of memory, affecting millions of individuals worldwide. With the dramatic increase in the prevalence of Alzheimer's disease, it is expected to impose extensive public health and economic burden. However, this burden is particularly heavy on the caregivers of Alzheimer's disease patients eliciting neuropsychiatric symptoms that include mood swings, hallucinations, and depression. Interestingly, these neuropsychiatric symptoms are shared across symptoms of bipolar disorder, schizophrenia, and major depression disorder. Despite the similarities in symptomatology, comorbidities of Alzheimer's disease and these neuropsychiatric disorders have not been studied in the Alzheimer's disease model. Here, we explore the comprehensive changes in gene expression of genes that are associated with bipolar disorder, schizophrenia, and major depression disorder through the microarray of an Alzheimer's disease animal model, the forebrain specific PSEN double knockout mouse. To analyze the genes related with these three neuropsychiatric disorders within the scope of our microarray data, we used selected 1207 of a total of 45,037 genes that satisfied our selection criteria. These genes were selected on the basis of 14 Gene Ontology terms significantly relevant with the three disorders which were identified by previous research conducted by the Psychiatric Genomics Consortium. Our study revealed that the forebrain specific deletion of Alzheimer's disease genes can significantly alter neuropsychiatric disorder associated genes. Most importantly, most of these significantly altered genes were found to be involved with schizophrenia. Taken together, we suggest that the synaptic dysfunction by mutation of Alzheimer's disease genes can lead to the manifestation of not only memory loss and impairments in cognition, but also neuropsychiatric symptoms.
Keywords: Alzheimer’s disease; big data; bipolar disorder; major depression disorder; microarray; neuropsychiatric symptoms; schizophrenia.
Figures


References
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

