In Search for Reliable Markers of Glioma-Induced Polarization of Microglia
- PMID: 29963047
- PMCID: PMC6013650
- DOI: 10.3389/fimmu.2018.01329
In Search for Reliable Markers of Glioma-Induced Polarization of Microglia
Abstract
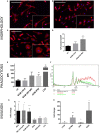
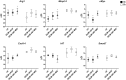
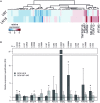
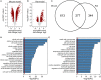
Immune cells accumulating in the microenvironment of malignant tumors are tumor educated and contribute to its growth, progression, and evasion of antitumor immune responses. Glioblastoma (GBM), the common and most malignant primary brain tumor in adults, shows considerable accumulation of resident microglia and peripheral macrophages, and their polarization into tumor-supporting cells. There are controversies regarding a functional phenotype of glioma-associated microglia/macrophages (GAMs) due to a lack of consistent markers. Previous categorization of GAM polarization toward the M2 phenotype has been found inaccurate because of oversimplification of highly complex and heterogeneous responses. In this study, we characterized functional responses and gene expression in mouse and human microglial cultures exposed to fresh conditioned media [glioma-conditioned medium (GCM)] from human U87 and LN18 glioma cells. Functional analyses revealed mutual communication reflected by strong stimulation of glioma invasion by microglial cells and increased microglial phagocytosis after GCM treatment. To define transcriptomic markers of GCM-activated microglia, we performed selected and global gene expression analyses of stimulated microglial cells. We found activated pathways associated with immune evasion and TGF signaling. We performed computational comparison of the expression patterns of GAMs from human GBMs and rodent experimental gliomas to select genes consistently changed in different datasets. The analyses of marker genes in GAMs from different experimental models and clinical samples revealed only a small set of common genes, which reflects variegated responses in clinical and experimental settings. Tgm2 and Gpnmb were the only two genes common in the analyzed data sets. We discuss potential sources of the observed differences and stress a great need for definitive elucidation of a functional state of GAMs.
Keywords: functional phenotype; glioma; glioma-associated microglia/macrophages; microglia; transcriptomics.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

