Bidirectional encoding of motion contrast in the mouse superior colliculus
- PMID: 29963987
- PMCID: PMC6050041
- DOI: 10.7554/eLife.35261
Bidirectional encoding of motion contrast in the mouse superior colliculus
Abstract
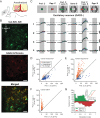
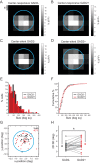
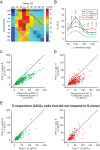
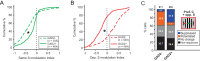
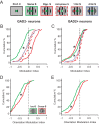
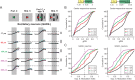
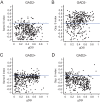
Detection of salient objects in the visual scene is a vital aspect of an animal's interactions with its environment. Here, we show that neurons in the mouse superior colliculus (SC) encode visual saliency by detecting motion contrast between stimulus center and surround. Excitatory neurons in the most superficial lamina of the SC are contextually modulated, monotonically increasing their response from suppression by the same-direction surround to maximal potentiation by an oppositely-moving surround. The degree of this potentiation declines with depth in the SC. Inhibitory neurons are suppressed by any surround at all depths. These response modulations in both neuronal populations are much more prominent to direction contrast than to phase, temporal frequency, or static orientation contrast, suggesting feature-specific saliency encoding in the mouse SC. Together, our findings provide evidence supporting locally generated feature representations in the SC, and lay the foundations towards a mechanistic and evolutionary understanding of their emergence.
Keywords: center-surround interactions; direction selectivity; mouse; neuroscience; saliency; superior colliculus; two-photon imaging; vision.
© 2018, Barchini et al.
Conflict of interest statement
JB, XS, HC, JC No competing interests declared
Figures






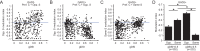
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

