A Radical Clock Probe Uncouples H Atom Abstraction from Thioether Cross-Link Formation by the Radical S-Adenosyl-l-methionine Enzyme SkfB
- PMID: 29965747
- PMCID: PMC6094349
- DOI: 10.1021/acs.biochem.8b00537
A Radical Clock Probe Uncouples H Atom Abstraction from Thioether Cross-Link Formation by the Radical S-Adenosyl-l-methionine Enzyme SkfB
Abstract
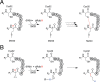
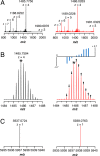
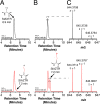
Sporulation killing factor (SKF) is a ribosomally synthesized and post-translationally modified peptide (RiPP) produced by Bacillus. SKF contains a thioether cross-link between the α-carbon at position 40 and the thiol of Cys32, introduced by a member of the radical S-adenosyl-l-methionine (SAM) superfamily, SkfB. Radical SAM enzymes employ a 4Fe-4S cluster to bind and reductively cleave SAM to generate a 5'-deoxyadenosyl radical. SkfB utilizes this radical intermediate to abstract the α-H atom at Met40 to initiate cross-linking. In addition to the cluster that binds SAM, SkfB also has an auxiliary cluster, the function of which is not known. We demonstrate that a substrate analogue with a cyclopropylglycine (CPG) moiety replacing the wild-type Met40 side chain forgoes thioether cross-linking for an alternative radical ring opening of the CPG side chain. The ring opening reaction also takes place with a catalytically inactive SkfB variant in which the auxiliary Fe-S cluster is absent. Therefore, the CPG-containing peptide uncouples H atom abstraction from thioether bond formation, limiting the role of the auxiliary cluster to promoting thioether cross-link formation. CPG proves to be a valuable tool for uncoupling H atom abstraction from peptide modification in RiPP maturases and demonstrates potential to leverage RS enzyme reactivity to create noncanonical amino acids.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

References
-
- Sofia H. J.; Chen G.; Hetzler B. G.; Reyes-Spindola J.F.; Miller N. E. (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29, 1097–1106. 10.1093/nar/29.5.1097. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

