RecQ helicases in the malaria parasite Plasmodium falciparum affect genome stability, gene expression patterns and DNA replication dynamics
- PMID: 29965959
- PMCID: PMC6044543
- DOI: 10.1371/journal.pgen.1007490
RecQ helicases in the malaria parasite Plasmodium falciparum affect genome stability, gene expression patterns and DNA replication dynamics
Abstract
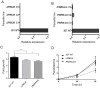
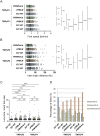
The malaria parasite Plasmodium falciparum has evolved an unusual genome structure. The majority of the genome is relatively stable, with mutation rates similar to most eukaryotic species. However, some regions are very unstable with high recombination rates, driving the generation of new immune evasion-associated var genes. The molecular factors controlling the inconsistent stability of this genome are not known. Here we studied the roles of the two putative RecQ helicases in P. falciparum, PfBLM and PfWRN. When PfWRN was knocked down, recombination rates increased four-fold, generating chromosomal abnormalities, a high rate of chimeric var genes and many microindels, particularly in known 'fragile sites'. This is the first identification of a gene involved in suppressing recombination and maintaining genome stability in Plasmodium. By contrast, no change in mutation rate appeared when the second RecQ helicase, PfBLM, was mutated. At the transcriptional level, however, both helicases evidently modulate the transcription of large cohorts of genes, with several hundred genes-including a large proportion of vars-showing deregulated expression in each RecQ mutant. Aberrant processing of stalled replication forks is a possible mechanism underlying elevated mutation rates and this was assessed by measuring DNA replication dynamics in the RecQ mutant lines. Replication forks moved slowly and stalled at elevated rates in both mutants, confirming that RecQ helicases are required for efficient DNA replication. Overall, this work identifies the Plasmodium RecQ helicases as major players in DNA replication, antigenic diversification and genome stability in the most lethal human malaria parasite, with important implications for genome evolution in this pathogen.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




Similar articles
-
Elucidation of DNA Repair Function of PfBlm and Potentiation of Artemisinin Action by a Small-Molecule Inhibitor of RecQ Helicase.mSphere. 2020 Nov 25;5(6):e00956-20. doi: 10.1128/mSphere.00956-20. mSphere. 2020. PMID: 33239368 Free PMC article.
-
Plasmodium falciparum Bloom homologue, a nucleocytoplasmic protein, translocates in 3' to 5' direction and is essential for parasite growth.Biochim Biophys Acta. 2016 May;1864(5):594-608. doi: 10.1016/j.bbapap.2016.02.016. Epub 2016 Feb 23. Biochim Biophys Acta. 2016. PMID: 26917473
-
DNA helicase RecQ1 regulates mutually exclusive expression of virulence genes in Plasmodium falciparum via heterochromatin alteration.Proc Natl Acad Sci U S A. 2019 Feb 19;116(8):3177-3182. doi: 10.1073/pnas.1811766116. Epub 2019 Feb 6. Proc Natl Acad Sci U S A. 2019. PMID: 30728298 Free PMC article.
-
The varieties of gene amplification, diversification and hypervariability in the human malaria parasite, Plasmodium falciparum.Mol Biochem Parasitol. 2009 Aug;166(2):109-16. doi: 10.1016/j.molbiopara.2009.04.003. Epub 2009 Apr 16. Mol Biochem Parasitol. 2009. PMID: 19375460 Review.
-
Functions of RecQ family helicases: possible involvement of Bloom's and Werner's syndrome gene products in guarding genome integrity during DNA replication.J Biochem. 2001 Apr;129(4):501-7. doi: 10.1093/oxfordjournals.jbchem.a002883. J Biochem. 2001. PMID: 11275547 Review.
Cited by
-
Discovery of RUF6 ncRNA-interacting proteins involved in P. falciparum immune evasion.Life Sci Alliance. 2022 Nov 15;6(1):e202201577. doi: 10.26508/lsa.202201577. Print 2023 Jan. Life Sci Alliance. 2022. PMID: 36379669 Free PMC article.
-
DNA replication dynamics are associated with genome composition in Plasmodium species.Nucleic Acids Res. 2025 Feb 8;53(4):gkaf111. doi: 10.1093/nar/gkaf111. Nucleic Acids Res. 2025. PMID: 39997219 Free PMC article.
-
An analysis of large structural variation in global Plasmodium falciparum isolates identifies a novel duplication of the chloroquine resistance associated gene.Sci Rep. 2019 Jun 4;9(1):8287. doi: 10.1038/s41598-019-44599-0. Sci Rep. 2019. PMID: 31164664 Free PMC article.
-
Host-parasite interactions during Plasmodium infection: Implications for immunotherapies.Front Immunol. 2023 Jan 4;13:1091961. doi: 10.3389/fimmu.2022.1091961. eCollection 2022. Front Immunol. 2023. PMID: 36685595 Free PMC article. Review.
-
Evolution of Host Specificity by Malaria Parasites through Altered Mechanisms Controlling Genome Maintenance.mBio. 2020 Mar 17;11(2):e03272-19. doi: 10.1128/mBio.03272-19. mBio. 2020. PMID: 32184256 Free PMC article.
References
-
- WHO. World Malaria Report 2016. WHO website. 2016. http://www.who.int/malaria/publications/world_malaria_report/en/.
-
- Guler JL, Freeman DL, Ahyong V, Patrapuvich R, White J, Gujjar R, et al. Asexual populations of the human malaria parasite, Plasmodium falciparum, use a two-step genomic strategy to acquire accurate, beneficial DNA amplifications. PLoS pathogens. 2013;9(5):e1003375 10.1371/journal.ppat.1003375 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

