The presence of genetic risk variants within PTPN2 and PTPN22 is associated with intestinal microbiota alterations in Swiss IBD cohort patients
- PMID: 29965986
- PMCID: PMC6028086
- DOI: 10.1371/journal.pone.0199664
The presence of genetic risk variants within PTPN2 and PTPN22 is associated with intestinal microbiota alterations in Swiss IBD cohort patients
Abstract
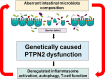
Background: Genetic risk factors, intestinal microbiota and a dysregulated immune system contribute to the pathogenesis of inflammatory bowel disease (IBD). We have previously demonstrated that dysfunction of protein tyrosine phosphatase non-receptor type 2 (PTPN2) and PTPN22 contributes to alterations of intestinal microbiota and the onset of chronic intestinal inflammation in vivo. Here, we investigated the influence of PTPN2 and PTPN22 gene variants on intestinal microbiota composition in IBD patients.
Methods: Bacterial DNA from mucosa-associated samples of 75 CD and 57 UC patients were sequenced using 16S rRNA sequencing approach. Microbial analysis, including alpha diversity, beta diversity and taxonomical analysis by comparing to PTPN2 (rs1893217) and PTPN22 (rs2476601) genotypes was performed in QIIME, the phyloseq R package and MaAsLin pipeline.
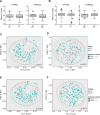
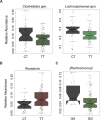
Results: In PTPN2 variant UC patients, we detected an increase in relative abundance of unassigned genera from Clostridiales and Lachnospiraceae families and reduction of Roseburia when compared to PTPN2 wild-type (WT) patients. Ruminoccocus was increased in PTPN22 variant UC patients. In CD patients with severe disease course, Faecalibacterium, Bilophila, Coprococcus, unclassified Erysipelotrichaeceae, unassigned genera from Clostridiales and Ruminococcaceae families were reduced and Bacteroides were increased in PTPN2 WT carriers, while Faecalibacterium, Bilophila, Coprococcus, and Erysipelotrichaeceae were reduced in PTPN22 WT patients when compared to patients with mild disease. In UC patients with severe disease, relative abundance of Lachnobacterium was reduced in PTPN2 and PTPN22 WT patients, Dorea was increased in samples from PTPN22 WT carriers and an unassigned genus from Ruminococcaceae gen. was increased in patients with PTPN2 variant genotype.
Conclusions: We identified that IBD-associated genetic risk variants, disease severity and the interaction of these factors are related to significant alterations in intestinal microbiota composition of IBD patients.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–73. doi: 10.1126/science.1223490 ; PubMed Central PMCID: PMC4420145. - DOI - PMC - PubMed
-
- Yilmaz B, Portugal S, Tran TM, Gozzelino R, Ramos S, Gomes J, et al. Gut microbiota elicits a protective immune response against malaria transmission. Cell. 2014;159(6):1277–89. doi: 10.1016/j.cell.2014.10.053 ; PubMed Central PMCID: PMC4261137. - DOI - PMC - PubMed
-
- Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–70. doi: 10.1016/j.cell.2012.01.035 ; PubMed Central PMCID: PMCPMC5050011. - DOI - PMC - PubMed
-
- Kaur N, Chen CC, Luther J, Kao JY. Intestinal dysbiosis in inflammatory bowel disease. Gut Microbes. 2011;2(4):211–6. doi: 10.4161/gmic.2.4.17863 . - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

