Atg9 proteins, not so different after all
- PMID: 29966469
- PMCID: PMC6103728
- DOI: 10.1080/15548627.2018.1477382
Atg9 proteins, not so different after all
Abstract
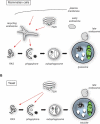
Macroautophagy (hereafter autophagy) is a catabolic pathway present in all eukaryotic cells. The yeast Saccharomyces cerevisiae has been pivotal in the identification and characterization of the key autophagy-related (Atg) proteins, which play a central role in the generation of autophagosomes. The components of the core Atg/ATG machinery and their functions are highly conserved among species, although mammalian cells also have isoforms and auxiliary factors. Atg9/ATG9 is the only transmembrane protein that is part of the core Atg/ATG machinery, but it appears to have divergent localizations and molecular roles in yeast and mammals. A recent experimental analysis of the yeast endo-lysosomal system by the laboratory of Benjamin Glick, however, suggests a more simple organization of this membrane system. Although this study has not examined yeast Atg9, its findings place this protein in the same compartments as its mammalian counterpart. Here, we will discuss the implications of this conceptual change on the trafficking of yeast Atg9 and its function in autophagy.
Keywords: ATG9A; Atg9; endosomes; recycling endosomes; trafficking; trans-Golgi network.
Figures
References
-
- Lamb CA, Yoshimori T, Tooze SA.. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14:759–774. - PubMed
-
- Mizushima N, Yoshimori T, Ohsumi Y.. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. - PubMed
-
- Young ARJ, Chan EYW, Hu XW, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci. 2006;119:3888–3900. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
