Slice-level diffusion encoding for motion and distortion correction
- PMID: 29966941
- PMCID: PMC6191883
- DOI: 10.1016/j.media.2018.06.008
Slice-level diffusion encoding for motion and distortion correction
Abstract
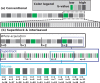
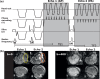
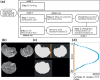
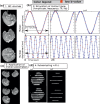
Advances in microstructural modelling are leading to growing requirements on diffusion MRI acquisitions, namely sensitivity to smaller structures and better resolution of the geometric orientations. The resulting acquisitions contain highly attenuated images that present particular challenges when there is motion and geometric distortion. This study proposes to address these challenges by breaking with the conventional one-volume-one-encoding paradigm employed in conventional diffusion imaging using single-shot Echo Planar Imaging. By enabling free choice of the diffusion encoding on the slice level, a higher temporal sampling of slices with low b-value can be achieved. These allow more robust motion correction, and in combination with a second reversed phase-encoded echo, also dynamic distortion correction. These proposed advances are validated on phantom and adult experiments and employed in a study of eight foetal subjects. Equivalence in obtained diffusion quantities with the conventional method is demonstrated as well as benefits in distortion and motion correction. The resulting capability can be combined with any acquisition parameters including multiband imaging and allows application to diffusion MRI studies in general.
Keywords: Diffusion; EPI; Foetal imaging; MRI; Microstructure; Motion correction.
Copyright © 2018 The Authors. Published by Elsevier B.V. All rights reserved.
Figures







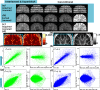
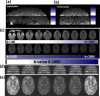
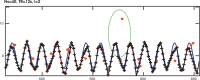
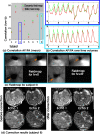
Similar articles
-
Simultaneous Motion and Distortion Correction Using Dual-Echo Diffusion-Weighted MRI.J Neuroimaging. 2020 May;30(3):276-285. doi: 10.1111/jon.12708. Epub 2020 May 6. J Neuroimaging. 2020. PMID: 32374453 Free PMC article.
-
High-resolution distortion-free diffusion imaging using hybrid spin-warp and echo-planar PSF-encoding approach.Neuroimage. 2017 Mar 1;148:20-30. doi: 10.1016/j.neuroimage.2017.01.008. Epub 2017 Jan 5. Neuroimage. 2017. PMID: 28065851
-
Single-shot echo planar time-resolved imaging for multi-echo functional MRI and distortion-free diffusion imaging.Magn Reson Med. 2025 Mar;93(3):993-1013. doi: 10.1002/mrm.30327. Epub 2024 Oct 20. Magn Reson Med. 2025. PMID: 39428674 Free PMC article.
-
Image formation in diffusion MRI: A review of recent technical developments.J Magn Reson Imaging. 2017 Sep;46(3):646-662. doi: 10.1002/jmri.25664. Epub 2017 Feb 14. J Magn Reson Imaging. 2017. PMID: 28194821 Free PMC article. Review.
-
Retrospective motion correction in foetal MRI for clinical applications: existing methods, applications and integration into clinical practice.Br J Radiol. 2023 Jul;96(1147):20220071. doi: 10.1259/bjr.20220071. Epub 2022 Aug 8. Br J Radiol. 2023. PMID: 35834425 Free PMC article. Review.
Cited by
-
Fetal Echoplanar Imaging: Promises and Challenges.Top Magn Reson Imaging. 2019 Oct;28(5):245-254. doi: 10.1097/RMR.0000000000000219. Top Magn Reson Imaging. 2019. PMID: 31592991 Free PMC article. Review.
-
HAITCH: A framework for distortion and motion correction in fetal multi-shell diffusion-weighted MRI.Imaging Neurosci (Camb). 2025 Feb 26;3:imag_a_00490. doi: 10.1162/imag_a_00490. eCollection 2025. Imaging Neurosci (Camb). 2025. PMID: 40800816 Free PMC article.
-
Combined diffusion-relaxometry microstructure imaging: Current status and future prospects.Magn Reson Med. 2021 Dec;86(6):2987-3011. doi: 10.1002/mrm.28963. Epub 2021 Aug 19. Magn Reson Med. 2021. PMID: 34411331 Free PMC article. Review.
-
Prenatal exposure to air pollution is associated with structural changes in the neonatal brain.Environ Int. 2023 Apr;174:107921. doi: 10.1016/j.envint.2023.107921. Epub 2023 Apr 9. Environ Int. 2023. PMID: 37058974 Free PMC article.
-
Spatiotemporal tissue maturation of thalamocortical pathways in the human fetal brain.Elife. 2023 Apr 3;12:e83727. doi: 10.7554/eLife.83727. Elife. 2023. PMID: 37010273 Free PMC article.
References
-
- Alexander D.C., Hubbard P.L., Hall M.G., Moore E.A., Ptito M., Parker G.J., Dyrby T.B. Orientationally invariant indices of axon diameter and density from diffusion MRI. Neuroimage. 2010;52(4):1374–1389. - PubMed
-
- Andersson J.L.R., Skare S., Ashburner J. How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage. 2003;20(2):870–888. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

