Minor zygotic gene activation is essential for mouse preimplantation development
- PMID: 29967139
- PMCID: PMC6055165
- DOI: 10.1073/pnas.1804309115
Minor zygotic gene activation is essential for mouse preimplantation development
Abstract
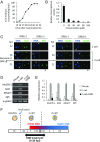
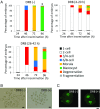
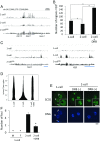
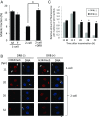
In mice, transcription initiates at the mid-one-cell stage and transcriptional activity dramatically increases during the two-cell stage, a process called zygotic gene activation (ZGA). Associated with ZGA is a marked change in the pattern of gene expression that occurs after the second round of DNA replication. To distinguish ZGA before and after the second-round DNA replication, the former and latter are called minor and major ZGA, respectively. Although major ZGA are required for development beyond the two-cell stage, the function of minor ZGA is not well understood. Transiently inhibiting minor ZGA with 5, 6-dichloro-1-β-d-ribofuranosyl-benzimidazole (DRB) resulted in the majority of embryos arresting at the two-cell stage and retention of the H3K4me3 mark that normally decreases. After release from DRB, at which time major ZGA normally occurred, transcription initiated with characteristics of minor ZGA but not major ZGA, although degradation of maternal mRNA normally occurred. Thus, ZGA occurs sequentially starting with minor ZGA that is critical for the maternal-to-zygotic transition.
Keywords: gene expression; maternal-to-zygotic transition; minor zygotic gene activation; preimplantation mouse embryo.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Bleil JD, Wassarman PM. Mammalian sperm-egg interaction: Identification of a glycoprotein in mouse egg zonae pellucidae possessing receptor activity for sperm. Cell. 1980;20:873–882. - PubMed
-
- McGrath SA, Esquela AF, Lee SJ. Oocyte-specific expression of growth/differentiation factor-9. Mol Endocrinol. 1995;9:131–136. - PubMed
-
- Rajkovic A, Matzuk MM. Functional analysis of oocyte-expressed genes using transgenic models. Mol Cell Endocrinol. 2002;187:5–9. - PubMed
-
- Moore GPM, Lintern-Moore S. A correlation between growth and RNA synthesis in the mouse oocyte. J Reprod Fertil. 1974;39:163–166. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

