Chromatin organization by an interplay of loop extrusion and compartmental segregation
- PMID: 29967174
- PMCID: PMC6055145
- DOI: 10.1073/pnas.1717730115
Chromatin organization by an interplay of loop extrusion and compartmental segregation
Abstract
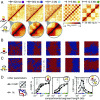
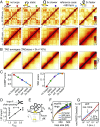
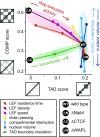
Mammalian chromatin is spatially organized at many scales showing two prominent features in interphase: (i) alternating regions (1-10 Mb) of active and inactive chromatin that spatially segregate into different compartments, and (ii) domains (<1 Mb), that is, regions that preferentially interact internally [topologically associating domains (TADs)] and are central to gene regulation. There is growing evidence that TADs are formed by active extrusion of chromatin loops by cohesin, whereas compartmentalization is established according to local chromatin states. Here, we use polymer simulations to examine how loop extrusion and compartmental segregation work collectively and potentially interfere in shaping global chromosome organization. A model with differential attraction between euchromatin and heterochromatin leads to phase separation and reproduces compartmentalization as observed in Hi-C. Loop extrusion, essential for TAD formation, in turn, interferes with compartmentalization. Our integrated model faithfully reproduces Hi-C data from puzzling experimental observations where altering loop extrusion also led to changes in compartmentalization. Specifically, depletion of chromatin-associated cohesin reduced TADs and revealed finer compartments, while increased processivity of cohesin strengthened large TADs and reduced compartmentalization; and depletion of the TAD boundary protein CTCF weakened TADs while leaving compartments unaffected. We reveal that these experimental perturbations are special cases of a general polymer phenomenon of active mixing by loop extrusion. Our results suggest that chromatin organization on the megabase scale emerges from competition of nonequilibrium active loop extrusion and epigenetically defined compartment structure.
Keywords: Hi-C; active matter; chromatin; genome architecture; polymer physics.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. - PubMed
-
- Bonev B, Cavalli G. Organization and function of the 3D genome. Nat Rev Genet. 2016;17:661–678. - PubMed
-
- Osborne CS, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

