Shedding of CD16 disassembles the NK cell immune synapse and boosts serial engagement of target cells
- PMID: 29967280
- PMCID: PMC6122987
- DOI: 10.1083/jcb.201712085
Shedding of CD16 disassembles the NK cell immune synapse and boosts serial engagement of target cells
Abstract
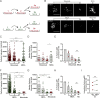
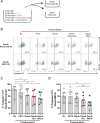
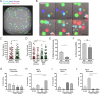
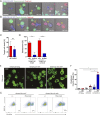
Natural Killer (NK) cells can engage multiple virally infected or tumor cells sequentially and deliver perforin for cytolytic killing of these targets. Using microscopy to visualize degranulation from individual NK cells, we found that repeated activation via the Fc receptor CD16 decreased the amount of perforin secreted. However, perforin secretion was restored upon subsequent activation via a different activating receptor, NKG2D. Repeated stimulation via NKG2D also decreased perforin secretion, but this was not rescued by stimulation via CD16. These different outcomes of sequential stimulation could be accounted for by shedding of CD16 being triggered by cellular activation. The use of pharmacological inhibitors and NK cells transfected to express a noncleavable form of CD16 revealed that CD16 shedding also increased NK cell motility and facilitated detachment of NK cells from target cells. Disassembly of the immune synapse caused by CD16 shedding aided NK cell survival and boosted serial engagement of target cells. Thus, counterintuitively, shedding of CD16 may positively impact immune responses.
© 2018 Srpan et al.
Figures




Comment in
-
Natural killers shed attachments to kill again.J Cell Biol. 2018 Sep 3;217(9):2983-2985. doi: 10.1083/jcb.201807105. Epub 2018 Aug 14. J Cell Biol. 2018. PMID: 30108125 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

