Prenatal immune activation alters the adult neural epigenome but can be partly stabilised by a n-3 polyunsaturated fatty acid diet
- PMID: 29967385
- PMCID: PMC6028639
- DOI: 10.1038/s41398-018-0167-x
Prenatal immune activation alters the adult neural epigenome but can be partly stabilised by a n-3 polyunsaturated fatty acid diet
Abstract
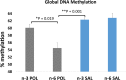
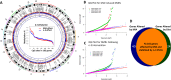
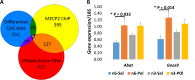
An unstable epigenome is implicated in the pathophysiology of neurodevelopmental disorders such as schizophrenia and autism. This is important because the epigenome is potentially modifiable. We have previously reported that adult offspring exposed to maternal immune activation (MIA) prenatally have significant global DNA hypomethylation in the hypothalamus. However, what genes had altered methylation state, their functional effects on gene expression and whether these changes can be moderated, have not been addressed. In this study, we used next-generation sequencing (NGS) for methylome profiling in a MIA rodent model of neurodevelopmental disorders. We assessed whether differentially methylated regions (DMRs) affected the chromatin state by mapping known DNase I hypersensitivity sites (DHSs), and selected overlapping genes to confirm a functional effect of MIA on gene expression using qPCR. Finally, we tested whether methylation differences elicited by MIA could be limited by post-natal dietary (omega) n-3 polyunsaturated fatty acid (PUFA) supplementation. These experiments were conducted using hypothalamic brain tissue from 12-week-old offspring of mice injected with viral analogue PolyI:C on gestation day 9 of pregnancy or saline on gestation day 9. Half of the animals from each group were fed a diet enriched with n-3 PUFA from weaning (MIA group, n = 12 units, n = 39 mice; Control group, n = 12 units, n = 38 mice). The results confirmed our previous finding that adult offspring exposed to MIA prenatally had significant global DNA hypomethylation. Furthermore, genes linked to synaptic plasticity were over-represented among differentially methylated genes following MIA. More than 80% of MIA-induced hypomethylated sites, including those affecting chromatin state and MECP2 binding, were stabilised by the n-3 PUFA intervention. MIA resulted in increased expression of two of the 'top five' genes identified from an integrated analysis of DMRs, DHSs and MECP2 binding sites, namely Abat (t = 2.46, p < 0.02) and Gnas9 (t = 2.96, p < 0.01), although these changes were not stabilised by dietary intervention. Thus, prenatal MIA exposure impacts upon the epigenomic regulation of gene pathways linked to neurodevelopmental conditions; and many of the changes can be attenuated by a low-cost dietary intervention.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

