Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4
- PMID: 29967539
- PMCID: PMC6044470
- DOI: 10.1038/s41594-018-0085-x
Structure of the Cdc48 ATPase with its ubiquitin-binding cofactor Ufd1-Npl4
Abstract
Many polyubiquitinated proteins are extracted from membranes or complexes by the conserved ATPase Cdc48 (in yeast; p97 or VCP in mammals) before proteasomal degradation. Each Cdc48 hexamer contains two stacked ATPase rings (D1 and D2) and six N-terminal (N) domains. Cdc48 binds various cofactors, including the Ufd1-Npl4 heterodimer. Here, we report structures of the Cdc48-Ufd1-Npl4 complex from Chaetomium thermophilum. Npl4 interacts through its UBX-like domain with a Cdc48 N domain, and it uses two Zn2+-finger domains to anchor the enzymatically inactive Mpr1-Pad1 N-terminal (MPN) domain, homologous to domains found in several isopeptidases, to the top of the D1 ATPase ring. The MPN domain of Npl4 is located above Cdc48's central pore, a position similar to the MPN domain from deubiquitinase Rpn11 in the proteasome. Our results indicate that Npl4 is unique among Cdc48 cofactors and suggest a mechanism for binding and translocation of polyubiquitinated substrates into the ATPase.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



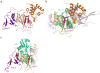
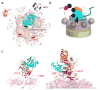
References
-
- Zhang X, et al. Structure of the AAA ATPase p97. Mol Cell. 2000;6:1473–1484. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

