Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience
- PMID: 29967940
- PMCID: PMC6060790
- DOI: 10.1007/s00401-018-1879-y
Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: the Heidelberg experience
Abstract
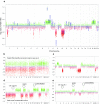
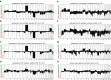
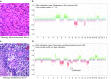
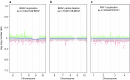
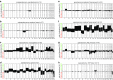
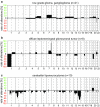
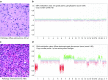
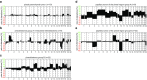
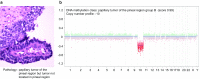
Recently, we described a machine learning approach for classification of central nervous system tumors based on the analysis of genome-wide DNA methylation patterns [6]. Here, we report on DNA methylation-based central nervous system (CNS) tumor diagnostics conducted in our institution between the years 2015 and 2018. In this period, more than 1000 tumors from the neurosurgical departments in Heidelberg and Mannheim and more than 1000 tumors referred from external institutions were subjected to DNA methylation analysis for diagnostic purposes. We describe our current approach to the integrated diagnosis of CNS tumors with a focus on constellations with conflicts between morphological and molecular genetic findings. We further describe the benefit of integrating DNA copy-number alterations into diagnostic considerations and provide a catalog of copy-number changes for individual DNA methylation classes. We also point to several pitfalls accompanying the diagnostic implementation of DNA methylation profiling and give practical suggestions for recurring diagnostic scenarios.
Keywords: Copy-number variation; DNA methylation; EPIC array; Tumor classification.
Figures




References
-
- Bady P, Sciuscio D, Diserens AC, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol. 2012;124:547–560. doi: 10.1007/s00401-012-1016-2. - DOI - PMC - PubMed
-
- Capper D, Engel NW, Stichel D et al (2018) DNA methylation-based reclassification of olfactory neuroblastoma. Acta Neuropathol. 10.1007/s00401-018-1854-7 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

