Crystal structure of the spliceosomal DEAH-box ATPase Prp2
- PMID: 29968674
- PMCID: PMC6038383
- DOI: 10.1107/S2059798318006356
Crystal structure of the spliceosomal DEAH-box ATPase Prp2
Abstract
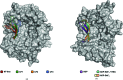
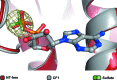
The DEAH-box ATPase Prp2 plays a key role in the activation of the spliceosome as it promotes the transition from the Bact to the catalytically active B* spliceosome. Here, four crystal structures of Prp2 are reported: one of the nucleotide-free state and three different structures of the ADP-bound state. The overall conformation of the helicase core, formed by two RecA-like domains, does not differ significantly between the ADP-bound and the nucleotide-free states. However, intrinsic flexibility of Prp2 is observed, varying the position of the C-terminal domains with respect to the RecA domains. Additionally, in one of the structures a unique ADP conformation is found which has not been observed in any other DEAH-box, DEAD-box or NS3/NPH-II helicase.
Keywords: DEAH-box; Prp2; RNA helicase; spliceosome.
open access.
Figures


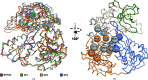
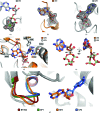
Similar articles
-
Crystal structure of Prp16 in complex with ADP.Acta Crystallogr F Struct Biol Commun. 2023 Aug 1;79(Pt 8):200-207. doi: 10.1107/S2053230X23005721. Epub 2023 Jul 25. Acta Crystallogr F Struct Biol Commun. 2023. PMID: 37548918 Free PMC article.
-
The structure of Prp2 bound to RNA and ADP-BeF3- reveals structural features important for RNA unwinding by DEAH-box ATPases.Acta Crystallogr D Struct Biol. 2021 Apr 1;77(Pt 4):496-509. doi: 10.1107/S2059798321001194. Epub 2021 Mar 30. Acta Crystallogr D Struct Biol. 2021. PMID: 33825710 Free PMC article.
-
Structural basis for RNA translocation by DEAH-box ATPases.Nucleic Acids Res. 2019 May 7;47(8):4349-4362. doi: 10.1093/nar/gkz150. Nucleic Acids Res. 2019. PMID: 30828714 Free PMC article.
-
Structure and function of spliceosomal DEAH-box ATPases.Biol Chem. 2023 Jul 17;404(8-9):851-866. doi: 10.1515/hsz-2023-0157. Print 2023 Jul 26. Biol Chem. 2023. PMID: 37441768 Review.
-
The DEAD-box helicase eIF4A: paradigm or the odd one out?RNA Biol. 2013 Jan;10(1):19-32. doi: 10.4161/rna.21966. Epub 2012 Sep 20. RNA Biol. 2013. PMID: 22995829 Free PMC article. Review.
Cited by
-
Auxiliary domains of the HrpB bacterial DExH-box helicase shape its RNA preferences.RNA Biol. 2020 May;17(5):637-650. doi: 10.1080/15476286.2020.1720376. Epub 2020 Feb 12. RNA Biol. 2020. PMID: 32050838 Free PMC article.
-
The RNA helicase DHX16 recognizes specific viral RNA to trigger RIG-I-dependent innate antiviral immunity.Cell Rep. 2022 Mar 8;38(10):110434. doi: 10.1016/j.celrep.2022.110434. Cell Rep. 2022. PMID: 35263596 Free PMC article.
-
Crystal structures of the DExH-box RNA helicase DHX9.Acta Crystallogr D Struct Biol. 2023 Nov 1;79(Pt 11):980-991. doi: 10.1107/S2059798323007611. Epub 2023 Oct 20. Acta Crystallogr D Struct Biol. 2023. PMID: 37860960 Free PMC article.
-
Molecular mechanism of the RNA helicase DHX37 and its activation by UTP14A in ribosome biogenesis.RNA. 2019 Jun;25(6):685-701. doi: 10.1261/rna.069609.118. Epub 2019 Mar 25. RNA. 2019. PMID: 30910870 Free PMC article.
-
Continuous millisecond conformational cycle of a DEAH box helicase reveals control of domain motions by atomic-scale transitions.Commun Biol. 2023 Apr 7;6(1):379. doi: 10.1038/s42003-023-04751-z. Commun Biol. 2023. PMID: 37029280 Free PMC article.
References
-
- Absmeier, E., Rosenberger, L., Apelt, L., Becke, C., Santos, K. F., Stelzl, U. & Wahl, M. C. (2015). Acta Cryst. D71, 762–771. - PubMed
-
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221. - PubMed
-
- Amlacher, S., Sarges, P., Flemming, D., van Noort, V., Kunze, R., Devos, D. P., Arumugam, M., Bork, P. & Hurt, E. (2011). Cell, 146, 277–289. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

