Trapping of the transport-segment DNA by the ATPase domains of a type II topoisomerase
- PMID: 29968711
- PMCID: PMC6030046
- DOI: 10.1038/s41467-018-05005-x
Trapping of the transport-segment DNA by the ATPase domains of a type II topoisomerase
Abstract
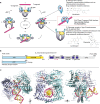
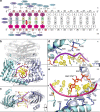
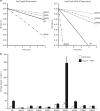
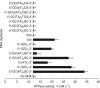
Type II topoisomerases alter DNA topology to control DNA supercoiling and chromosome segregation and are targets of clinically important anti-infective and anticancer therapeutics. They act as ATP-operated clamps to trap a DNA helix and transport it through a transient break in a second DNA. Here, we present the first X-ray crystal structure solved at 2.83 Å of a closed clamp complete with trapped T-segment DNA obtained by co-crystallizing the ATPase domain of S. pneumoniae topoisomerase IV with a nonhydrolyzable ATP analogue and 14-mer duplex DNA. The ATPase dimer forms a 22 Å protein hole occupied by the kinked DNA bound asymmetrically through positively charged residues lining the hole, and whose mutagenesis impacts the DNA decatenation, DNA relaxation and DNA-dependent ATPase activities of topo IV. These results and a side-bound DNA-ParE structure help explain how the T-segment DNA is captured and transported by a type II topoisomerase, and reveal a new enzyme-DNA interface for drug discovery.
Conflict of interest statement
The authors declare no competing interests.
Figures
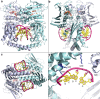


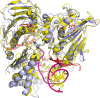
Similar articles
-
What makes a type IIA topoisomerase a gyrase or a Topo IV?Nucleic Acids Res. 2021 Jun 21;49(11):6027-6042. doi: 10.1093/nar/gkab270. Nucleic Acids Res. 2021. PMID: 33905522 Free PMC article. Review.
-
Structure of an 'open' clamp type II topoisomerase-DNA complex provides a mechanism for DNA capture and transport.Nucleic Acids Res. 2013 Nov;41(21):9911-23. doi: 10.1093/nar/gkt749. Epub 2013 Aug 21. Nucleic Acids Res. 2013. PMID: 23965305 Free PMC article.
-
ATP-bound conformation of topoisomerase IV: a possible target for quinolones in Streptococcus pneumoniae.J Bacteriol. 2003 Oct;185(20):6137-46. doi: 10.1128/JB.185.20.6137-6146.2003. J Bacteriol. 2003. PMID: 14526026 Free PMC article.
-
Breakage-reunion domain of Streptococcus pneumoniae topoisomerase IV: crystal structure of a gram-positive quinolone target.PLoS One. 2007 Mar 21;2(3):e301. doi: 10.1371/journal.pone.0000301. PLoS One. 2007. PMID: 17375187 Free PMC article.
-
The mechanism of negative DNA supercoiling: a cascade of DNA-induced conformational changes prepares gyrase for strand passage.DNA Repair (Amst). 2014 Apr;16:23-34. doi: 10.1016/j.dnarep.2014.01.011. Epub 2014 Feb 22. DNA Repair (Amst). 2014. PMID: 24674625 Review.
Cited by
-
What makes a type IIA topoisomerase a gyrase or a Topo IV?Nucleic Acids Res. 2021 Jun 21;49(11):6027-6042. doi: 10.1093/nar/gkab270. Nucleic Acids Res. 2021. PMID: 33905522 Free PMC article. Review.
-
Role of unique loops in oligomerization and ATPase function of Plasmodium falciparum gyrase B.Protein Sci. 2022 Feb;31(2):323-332. doi: 10.1002/pro.4217. Epub 2021 Nov 6. Protein Sci. 2022. PMID: 34716632 Free PMC article.
-
Gyrase and Topoisomerase IV: Recycling Old Targets for New Antibacterials to Combat Fluoroquinolone Resistance.ACS Infect Dis. 2024 Apr 12;10(4):1097-1115. doi: 10.1021/acsinfecdis.4c00128. Epub 2024 Apr 2. ACS Infect Dis. 2024. PMID: 38564341 Free PMC article. Review.
-
Structure-function analysis of the ATPase domain of African swine fever virus topoisomerase.mBio. 2024 Apr 10;15(4):e0308623. doi: 10.1128/mbio.03086-23. Epub 2024 Feb 27. mBio. 2024. PMID: 38411066 Free PMC article.
-
Unveiling the interdomain dynamics of type II DNA topoisomerase through all-atom simulations: Implications for understanding its catalytic cycle.Comput Struct Biotechnol J. 2023 Jul 22;21:3746-3759. doi: 10.1016/j.csbj.2023.07.019. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37602233 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

