Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design
- PMID: 29969259
- PMCID: PMC6334293
- DOI: 10.1021/acs.jmedchem.8b00040
Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design
Abstract
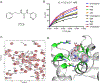
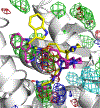
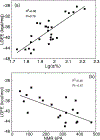
Protein-protein interactions (PPI) between the transcriptional repressor B-cell lymphoma 6 (BCL6) BTB domain (BCL6BTB) and its corepressors have emerged as a promising target for anticancer therapeutics. However, identification of potent, drug-like inhibitors of BCL6BTB has remained challenging. Using NMR-based screening of a library of fragment-like small molecules, we have identified a thiourea compound (7CC5) that binds to BCL6BTB. From this hit, the application of computer-aided drug design (CADD), medicinal chemistry, NMR spectroscopy, and X-ray crystallography has yielded an inhibitor, 15f, that demonstrated over 100-fold improved potency for BCL6BTB. This gain in potency was achieved by a unique binding mode that mimics the binding mode of the corepressor SMRT in the aromatic and the HDCH sites. The structure-activity relationship based on these new inhibitors will have a significant impact on the rational design of novel BCL6 inhibitors, facilitating the identification of therapeutics for the treatment of BCL6-dependent tumors.
Figures


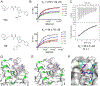


Similar articles
-
Progress toward B-Cell Lymphoma 6 BTB Domain Inhibitors for the Treatment of Diffuse Large B-Cell Lymphoma and Beyond.J Med Chem. 2021 Apr 22;64(8):4333-4358. doi: 10.1021/acs.jmedchem.0c01686. Epub 2021 Apr 12. J Med Chem. 2021. PMID: 33844535 Free PMC article.
-
Discovery of an Irreversible and Cell-Active BCL6 Inhibitor Selectively Targeting Cys53 Located at the Protein-Protein Interaction Interface.Biochemistry. 2018 Feb 27;57(8):1369-1379. doi: 10.1021/acs.biochem.7b00732. Epub 2018 Feb 7. Biochemistry. 2018. PMID: 29293322
-
Structural basis of Apt48 inhibition of the BCL6 BTB domain.Structure. 2022 Mar 3;30(3):396-407.e3. doi: 10.1016/j.str.2021.10.010. Epub 2021 Nov 12. Structure. 2022. PMID: 34774129
-
Therapeutic targeting of the BCL6 oncogene for diffuse large B-cell lymphomas.Leuk Lymphoma. 2008 May;49(5):874-82. doi: 10.1080/10428190801895345. Leuk Lymphoma. 2008. PMID: 18452090 Free PMC article. Review.
-
BCL6 as a therapeutic target for lymphoma.Expert Opin Ther Targets. 2018 Feb;22(2):143-152. doi: 10.1080/14728222.2018.1420782. Epub 2018 Jan 4. Expert Opin Ther Targets. 2018. PMID: 29262721 Review.
Cited by
-
Increased slow dynamics defines ligandability of BTB domains.Nat Commun. 2022 Nov 16;13(1):6989. doi: 10.1038/s41467-022-34599-6. Nat Commun. 2022. PMID: 36384931 Free PMC article.
-
Integrating Protein Interaction Surface Prediction with a Fragment-Based Drug Design: Automatic Design of New Leads with Fragments on Energy Surfaces.J Chem Inf Model. 2023 Jan 9;63(1):343-353. doi: 10.1021/acs.jcim.2c01408. Epub 2022 Dec 27. J Chem Inf Model. 2023. PMID: 36574607 Free PMC article.
-
Exploring protein-protein interactions using the site-identification by ligand competitive saturation methodology.Proteins. 2019 Apr;87(4):289-301. doi: 10.1002/prot.25650. Epub 2019 Jan 10. Proteins. 2019. PMID: 30582220 Free PMC article.
-
Progress toward B-Cell Lymphoma 6 BTB Domain Inhibitors for the Treatment of Diffuse Large B-Cell Lymphoma and Beyond.J Med Chem. 2021 Apr 22;64(8):4333-4358. doi: 10.1021/acs.jmedchem.0c01686. Epub 2021 Apr 12. J Med Chem. 2021. PMID: 33844535 Free PMC article.
-
Structural analysis of the PATZ1 BTB domain homodimer.Acta Crystallogr D Struct Biol. 2020 Jun 1;76(Pt 6):581-593. doi: 10.1107/S2059798320005355. Epub 2020 May 29. Acta Crystallogr D Struct Biol. 2020. PMID: 32496219 Free PMC article.
References
-
- Ye BH; Cattoretti G; Shen Q; Zhang J; Hawe N; de Waard R; Leung C; Nouri-Shirazi M; Orazi A; Chaganti RS; Rothman P; Stall AM; Pandolfi PP; Dalla-Favera R The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nature genet 1997, 16, 161–170. - PubMed
-
- Duy C; Hurtz C; Shojaee S; Cerchietti L; Geng H; Swaminathan S; Klemm L; Kweon SM; Nahar R; Braig M; Park E; Kim YM; Hofmann WK; Herzog S; Jumaa H; Koeffler HP; Yu JJ; Heisterkamp N; Graeber TG; Wu H; Ye BH; Melnick A; Muschen M BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature 2011, 473, 384–388. - PMC - PubMed
-
- Geng H; Brennan S; Milne TA; Chen WY; Li Y; Hurtz C; Kweon SM; Zickl L; Shojaee S; Neuberg D; Huang C; Biswas D; Xin Y; Racevskis J; Ketterling RP; Luger SM; Lazarus H; Tallman MS; Rowe JM; Litzow MR; Guzman ML; Allis CD; Roeder RG; Muschen M; Paietta E; Elemento O; Melnick AM Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer discov 2012, 2, 1004–1023. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

