Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks
- PMID: 29970003
- PMCID: PMC6029131
- DOI: 10.1186/s12864-018-4889-1
Prediction of RNA-protein sequence and structure binding preferences using deep convolutional and recurrent neural networks
Abstract
Background: RNA regulation is significantly dependent on its binding protein partner, known as the RNA-binding proteins (RBPs). Unfortunately, the binding preferences for most RBPs are still not well characterized. Interdependencies between sequence and secondary structure specificities is challenging for both predicting RBP binding sites and accurate sequence and structure motifs detection.
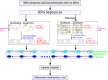
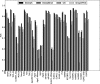
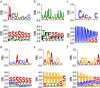
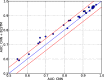
Results: In this study, we propose a deep learning-based method, iDeepS, to simultaneously identify the binding sequence and structure motifs from RNA sequences using convolutional neural networks (CNNs) and a bidirectional long short term memory network (BLSTM). We first perform one-hot encoding for both the sequence and predicted secondary structure, to enable subsequent convolution operations. To reveal the hidden binding knowledge from the observed sequences, the CNNs are applied to learn the abstract features. Considering the close relationship between sequence and predicted structures, we use the BLSTM to capture possible long range dependencies between binding sequence and structure motifs identified by the CNNs. Finally, the learned weighted representations are fed into a classification layer to predict the RBP binding sites. We evaluated iDeepS on verified RBP binding sites derived from large-scale representative CLIP-seq datasets. The results demonstrate that iDeepS can reliably predict the RBP binding sites on RNAs, and outperforms the state-of-the-art methods. An important advantage compared to other methods is that iDeepS can automatically extract both binding sequence and structure motifs, which will improve our understanding of the mechanisms of binding specificities of RBPs.
Conclusion: Our study shows that the iDeepS method identifies the sequence and structure motifs to accurately predict RBP binding sites. iDeepS is available at https://github.com/xypan1232/iDeepS .
Keywords: Bidirectional long short term memory network; Convolutional neural network; RNA-binding protein; Sequence motifs; Structure motifs.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

