Proteomic Profiling of Microtubule Self-organization in M-phase
- PMID: 29970457
- PMCID: PMC6166669
- DOI: 10.1074/mcp.RA118.000745
Proteomic Profiling of Microtubule Self-organization in M-phase
Abstract
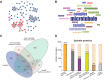
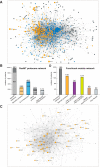
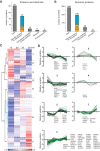
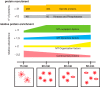
Microtubules (MTs) and associated proteins can self-organize into complex structures such as the bipolar spindle, a process in which RanGTP plays a major role. Addition of RanGTP to M-phase Xenopus egg extracts promotes the nucleation and self-organization of MTs into asters and bipolar-like structures in the absence of centrosomes or chromosomes. We show here that the complex proteome of these RanGTP-induced MT assemblies is similar to that of mitotic spindles. Using proteomic profiling we show that MT self-organization in the M-phase cytoplasm involves the non-linear and non-stoichiometric recruitment of proteins from specific functional groups. Our study provides for the first time a temporal understanding of the protein dynamics driving MT self-organization in M-phase.
Keywords: Cell biology; Cell division; Microtubule; Mitosis; Protein complex analysis; Protein-Protein Interactions; Proteomic profiling; RanGTP; Self-organization; Spindle.
© 2018 Rosas-Salvans et al.
Figures


References
-
- Karsenti E. (2008) Self-organization in cell biology: a brief history. Nature Reviews Mol. Cell Biol. 9, 255–262 - PubMed
-
- Vignaud T., Blanchoin L., and Thry M. (2012) Directed cytoskeleton self-organization. Trends Cell Biol. 22, 671–682 - PubMed
-
- Mitchison T., and Kirschner M. (1984) Dynamic instability of microtubule growth. Nature 312, 237–242 - PubMed
-
- Cortes S., Glade N., Chartier I., and Tabony J. (2006) Microtubule self-organisation by reaction-diffusion processes in miniature cell-sized containers and phospholipid vesicles. Biophys. Chem. 120, 168–177 - PubMed
-
- Surrey T., Nedelec F. J., Leibler S., and Karsenti E. (2001) Physical properties determining self-organization of motors and microtubules. Science 292, 1167–1171 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

