Quantitative Proteomic Analysis Provides Insights into Rice Defense Mechanisms against Magnaporthe oryzae
- PMID: 29970857
- PMCID: PMC6073306
- DOI: 10.3390/ijms19071950
Quantitative Proteomic Analysis Provides Insights into Rice Defense Mechanisms against Magnaporthe oryzae
Abstract
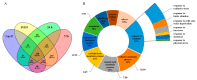
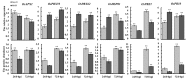
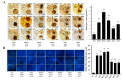
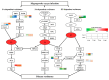
Blast disease is one of the major rice diseases, and causes nearly 30% annual yield loss worldwide. Resistance genes that have been cloned, however, are effective only against specific strains. In cultivation practice, broad-spectrum resistance to various strains is highly valuable, and requires researchers to investigate the basal defense responses that are effective for diverse types of pathogens. In this study, we took a quantitative proteomic approach and identified 634 rice proteins responsive to infections by both Magnaporthe oryzae strains Guy11 and JS153. These two strains have distinct pathogenesis mechanisms. Therefore, the common responding proteins represent conserved basal defense to a broad spectrum of blast pathogens. Gene ontology analysis indicates that the “responding to stimulus” biological process is explicitly enriched, among which the proteins responding to oxidative stress and biotic stress are the most prominent. These analyses led to the discoveries of OsPRX59 and OsPRX62 that are robust callose inducers, and OsHSP81 that is capable of inducing both ROS production and callose deposition. The identified rice proteins and biological processes may represent a conserved rice innate immune machinery that is of great value for breeding broad-spectrum resistant rice in the future.
Keywords: Magnaporthe oryzae; basal defense; iTRAQ; innate immunity; proteomics; rice blast.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

