Application of Docking Analysis in the Prediction and Biological Evaluation of the Lipoxygenase Inhibitory Action of Thiazolyl Derivatives of Mycophenolic Acid
- PMID: 29970872
- PMCID: PMC6099768
- DOI: 10.3390/molecules23071621
Application of Docking Analysis in the Prediction and Biological Evaluation of the Lipoxygenase Inhibitory Action of Thiazolyl Derivatives of Mycophenolic Acid
Abstract
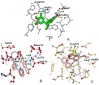
5-LOX inhibition is among the desired characteristics of anti-inflammatory drugs, while 15-LOX has also been considered as a drug target. Similarity in inhibition behavior between soybean LOX-1 and human 5-LOX has been observed and soybean LOX (sLOX) type 1b has been used for the evaluation of LOX inhibition in drug screening for years. After prediction of LOX inhibition by PASS and docking as well as toxicity by PROTOX and ToxPredict sixteen (E)-N-(thiazol-2-yl)-6-(4-hydroxy-6-methoxy-7-methyl-3-oxo-1,3-dihydroisobenzofuran-5-yl)-4-methylhex-4-enamide derivatives with lengths varying from about 15⁻20 Å were evaluated in vitro for LOX inhibitory action using the soybean lipoxygenase sLOX 1b. Docking analysis was performed using soybean LOX L-1 (1YGE), soybean LOX-3 (1JNQ), human 5-LOX (3O8Y and 3V99) and mammalian 15-LOX (1LOX) structures. Different dimensions of target center and docking boxes and a cavity prediction algorithm were used. The compounds exhibited inhibitory action between 2.5 μΜ and 165 μΜ. Substituents with an electronegative atom at two-bond proximity to position 4 of the thiazole led to enhanced activity. Docking results indicated that the LOX structures 1JNQ, 3V99 and 1LOX can effectively be used for estimation of LOX inhibition and amino acid interactions of these compounds.
Keywords: LOX; anti-inflammatory; docking; pharmacophore; thiazoles.
Conflict of interest statement
The authors declare no conflict of interest.
Figures










References
-
- Roberts L.J. Introduction: Lipids as regulators of cell function. Cell. Mol. Life Sci. 2002;59:727–728. doi: 10.1007/s00018-002-8461-3. - DOI
-
- Kuhn H. Lipoxygenases. In: Marks F., Fustenberger G., editors. Prostaglandins, Leukotrienes and Other Eicosanoids. Wiley-VCH; Weinheim, Germany: 1999. p. 109.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

