Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae
- PMID: 29970891
- PMCID: PMC6030107
- DOI: 10.1038/s41426-018-0124-z
Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae
Abstract
The rapid increase in carbapenem resistance among gram-negative bacteria has renewed focus on the importance of polymyxin antibiotics (colistin or polymyxin E). However, the recent emergence of plasmid-mediated colistin resistance determinants (mcr-1, -2, -3, -4, -5, -6, and -7), especially mcr-1, in carbapenem-resistant Enterobacteriaceae is a serious threat to global health. Here, we characterized a novel mobile colistin resistance gene, mcr-8, located on a transferrable 95,983-bp IncFII-type plasmid in Klebsiella pneumoniae. The deduced amino-acid sequence of MCR-8 showed 31.08%, 30.26%, 39.96%, 37.85%, 33.51%, 30.43%, and 37.46% identity to MCR-1, MCR-2, MCR-3, MCR-4, MCR-5, MCR-6, and MCR-7, respectively. Functional cloning indicated that the acquisition of the single mcr-8 gene significantly increased resistance to colistin in both Escherichia coli and K. pneumoniae. Notably, the coexistence of mcr-8 and the carbapenemase-encoding gene blaNDM was confirmed in K. pneumoniae isolates of livestock origin. Moreover, BLASTn analysis of mcr-8 revealed that this gene was present in a colistin- and carbapenem-resistant K. pneumoniae strain isolated from the sputum of a patient with pneumonia syndrome in the respiratory intensive care unit of a Chinese hospital in 2016. These findings indicated that mcr-8 has existed for some time and has disseminated among K. pneumoniae of both animal and human origin, further increasing the public health burden of antimicrobial resistance.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
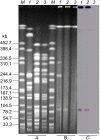
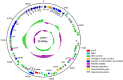
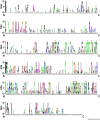


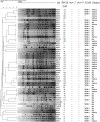

References
-
- Su, C. et al. Treatment outcome of non-carbapenemase-producing carbapenem-resistant Klebsiella pneumoniae infections: a multicenter study in Taiwan. Eur. J. Clin. Microbiol. Infect. Dis. 10.1007/s10096-017-3156-8 (2017). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
