Role of Optineurin in the Mitochondrial Dysfunction: Potential Implications in Neurodegenerative Diseases and Cancer
- PMID: 29971063
- PMCID: PMC6018216
- DOI: 10.3389/fimmu.2018.01243
Role of Optineurin in the Mitochondrial Dysfunction: Potential Implications in Neurodegenerative Diseases and Cancer
Abstract
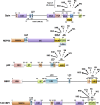
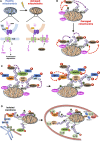
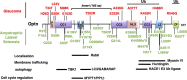
Optineurin (Optn) is a 577 aa protein encoded by the Optn gene. Mutations of Optn are associated with normal tension glaucoma and amyotrophic lateral sclerosis, and its gene has also been linked to the development of Paget's disease of bone and Crohn's disease. Optn is involved in diverse cellular functions, including NF-κB regulation, membrane trafficking, exocytosis, vesicle transport, reorganization of actin and microtubules, cell cycle control, and autophagy. Besides its role in xenophagy and autophagy of aggregates, Optn has been identified as a primary autophagy receptor, among the five adaptors that translocate to mitochondria during mitophagy. Mitophagy is a selective macroautophagy process during which irreparable mitochondria are degraded, preventing accumulation of defective mitochondria and limiting the release of reactive oxygen species and proapoptotic factors. Mitochondrial quality control via mitophagy is central to the health of cells. One of the important surveillance pathways of mitochondrial health is the recently defined signal transduction pathway involving the mitochondrial PTEN-induced putative kinase 1 (PINK1) protein and the cytosolic RING-between-RING ubiquitin ligase Parkin. Both of these proteins, when mutated, have been identified in certain forms of Parkinson's disease. By targeting ubiquitinated mitochondria to autophagosomes through its association with autophagy related proteins, Optn is responsible for a critical step in mitophagy. This review reports recent discoveries on the role of Optn in mitophagy and provides insight into its link with neurodegenerative diseases. We will also discuss the involvement of Optn in other pathologies in which mitophagy dysfunctions are involved including cancer.
Keywords: autophagy; autophagy receptor; cancer; mitophagy; neurodegenerative diseases; pathologies.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

