Genetic Architecture of Feeding Behavior and Feed Efficiency in a Duroc Pig Population
- PMID: 29971093
- PMCID: PMC6018414
- DOI: 10.3389/fgene.2018.00220
Genetic Architecture of Feeding Behavior and Feed Efficiency in a Duroc Pig Population
Abstract
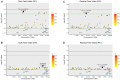
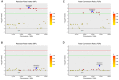
Increasing feed efficiency is a major goal of breeders as it can reduce production cost and energy consumption. However, the genetic architecture of feeding behavior and feed efficiency traits remains elusive. To investigate the genetic architecture of feed efficiency in pigs, three feeding behavior traits (daily feed intake, number of daily visits to feeder, and duration of each visit) and two feed efficiency traits (feed conversion ratio and residual feed intake) were considered. We performed genome-wide association studies (GWASs) of the five traits using a population of 1,008 Duroc pigs genotyped with an Illumina Porcine SNP50K BeadChip. A total of 9 genome-wide (P < 1.54E-06) and 35 suggestive (P < 3.08E-05) single nucleotide polymorphisms (SNPs) were detected. Two pleiotropic quantitative trait loci (QTLs) on SSC 1 and SSC 7 were found to affect more than one trait. Markers WU_10.2_7_18377044 and DRGA0001676 are two key SNPs for these two pleiotropic QTLs. Marker WU_10.2_7_18377044 on SSC 7 contributed 2.16 and 2.37% of the observed phenotypic variance for DFI and RFI, respectively. The other SNP DRGA0001676 on SSC 1 explained 3.22 and 5.46% of the observed phenotypic variance for FCR and RFI, respectively. Finally, functions of candidate genes and gene set enrichment analysis indicate that most of the significant pathways are associated with hormonal and digestive gland secretion during feeding. This study advances our understanding of the genetic mechanisms of feeding behavior and feed efficiency traits and provide an opportunity for increasing feeding efficiency using marker-assisted selection or genomic selection in pigs.
Keywords: Duroc; GWAS; feed conversion ratio; feed efficiency; feeding behavior; pigs; residual feed intake.
Figures


References
-
- Bartos P., Dolan A., Smutny L., Sistkova M., Celjak I., Soch M., et al. (2016). Effects of phytogenic feed additives on growth performance and on ammonia and greenhouse gases emissions in growing-finishing pigs. Anim. Feed Sci. Technol. 212 143–148. 10.1016/j.anifeedsci.2015.11.003 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

