A variant in LMX1A causes autosomal recessive severe-to-profound hearing impairment
- PMID: 29971487
- PMCID: PMC6094940
- DOI: 10.1007/s00439-018-1899-7
A variant in LMX1A causes autosomal recessive severe-to-profound hearing impairment
Abstract
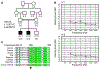
Hereditary hearing impairment is a common sensory disorder that is genetically and phenotypically heterogeneous. In this study, we used a homozygosity mapping and exome sequencing strategy to study a consanguineous Pakistani family with autosomal recessive severe-to-profound hearing impairment. This led to the identification of a missense variant (p.Ile369Thr) in the LMX1A gene affecting a conserved residue in the C-terminus of the protein, which was predicted damaging by an in silico bioinformatics analysis. The p.Ile369Thr variant disrupts several C-terminal and homeodomain residue interactions, including an interaction with homeodomain residue p.Val241 that was previously found to be involved in autosomal dominant progressive HI. LIM-homeodomain factor Lmx1a is expressed in the inner ear through development, shows a progressive restriction to non-sensory epithelia, and is important in the separation of the sensory and non-sensory domains in the inner ear. Homozygous Lmx1a mutant mice (Dreher) are deaf with dysmorphic ears with an abnormal morphogenesis and fused and misshapen sensory organs; however, computed tomography performed on a hearing-impaired family member did not reveal any cochleovestibular malformations. Our results suggest that LMX1A is involved in both human autosomal recessive and dominant sensorineural hearing impairment.
Keywords: Autosomal recessive hearing impairment; Deafness; Exome sequencing; LMX1A.
Conflict of interest statement
Compliance with ethical standards
Figures

References
-
- Alharazneh A, Luk L, Huth M et al. (2011) Functional hair cell mechanotransducer channels are required for aminoglycoside ototoxicity. PLoS One 6:e22347 https://doi.org/10.1371/journal.pone.00223 47 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

