Gut microbial diversity in two insectivorous bats: Insights into the effect of different sampling sources
- PMID: 29971963
- PMCID: PMC6530527
- DOI: 10.1002/mbo3.670
Gut microbial diversity in two insectivorous bats: Insights into the effect of different sampling sources
Abstract
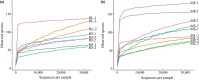
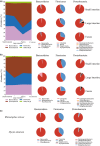
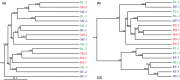
The gut microbiota is now known as a key factor in mammalian physiology and health. Our understanding of the gut microbial communities and their effects on ecology and evolution of their hosts is extremely limited in bats which represent the second largest mammalian order. In the current study, gut microbiota of three sampling sources (small intestine, large intestine, and feces) were characterized in two sympatric and insectivorous bats (Rhinolophus sinicus and Myotis altarium) by high-throughput sequencing of the V3-V4 region of the 16S rRNA gene. Combining with published studies, this work reveals that Gammaproteobacteria may be a dominant class in the whole Chiroptera and Fusobacteria is less observed in bats although it has been proven to be dominant in other mammals. Our results reveal that the sampling source influences alpha diversity of the microbial community in both studied species although no significant variations of beta diversity were observed, which support that fecal samples cannot be used as a proxy of the microbiota in other gut regions in wild animals.
Keywords: Myotis altarium; Rhinolophus sinicus; 16S rRNA; gut microbiota.
© 2018 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

