Multifunctional Activity-Based Protein Profiling of the Developing Lung
- PMID: 29972024
- PMCID: PMC6746415
- DOI: 10.1021/acs.jproteome.8b00086
Multifunctional Activity-Based Protein Profiling of the Developing Lung
Abstract
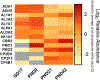
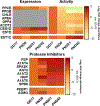
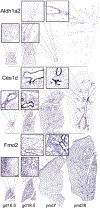
Lung diseases and disorders are a leading cause of death among infants. Many of these diseases and disorders are caused by premature birth and underdeveloped lungs. In addition to developmentally related disorders, the lungs are exposed to a variety of environmental contaminants and xenobiotics upon birth that can cause breathing issues and are progenitors of cancer. In order to gain a deeper understanding of the developing lung, we applied an activity-based chemoproteomics approach for the functional characterization of the xenometabolizing cytochrome P450 enzymes, active ATP and nucleotide binding enzymes, and serine hydrolases using a suite of activity-based probes (ABPs). We detected P450 activity primarily in the postnatal lung; using our ATP-ABP, we characterized a wide range of ATPases and other active nucleotide- and nucleic acid-binding enzymes involved in multiple facets of cellular metabolism throughout development. ATP-ABP targets include kinases, phosphatases, NAD- and FAD-dependent enzymes, RNA/DNA helicases, and others. The serine hydrolase-targeting probe detected changes in the activities of several proteases during the course of lung development, yielding insights into protein turnover at different stages of development. Select activity-based probe targets were then correlated with RNA in situ hybridization analyses of lung tissue sections.
Keywords: activity-based protein profiling; chemoproteomics; lung development.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
-
- Kochanek KD; Murphy SL; Xu JQ; Arias E Mortality in the United States, 2016; National Center for Health Statistics: Hyattsville, MD, 2017; NCHS Data Brief No. 293.
-
- Pond SM; Tozer TN First-Pass Elimination Basic Concepts and Clinical Consequences. Clin. Pharmacokinet 1984, 9 (1), 1–25. - PubMed
-
- Zhang JY; Wang Y; Prakash C Xenobiotic-metabolizing enzymes in human lung. Curr. Drug Metab 2006, 7 (8), 939–48. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

