A novel LRAT mutation affecting splicing in a family with early onset retinitis pigmentosa
- PMID: 29973277
- PMCID: PMC6033202
- DOI: 10.1186/s40246-018-0165-3
A novel LRAT mutation affecting splicing in a family with early onset retinitis pigmentosa
Abstract
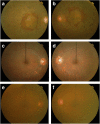
Background and purpose: Retinitis pigmentosa is an important cause of severe visual dysfunction. This study reports a novel splicing mutation in the lecithin retinol acyltransferase (LRAT) gene associated with early onset retinitis pigmentosa and characterizes the effects of this mutation on mRNA splicing and structure.
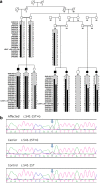
Methods: Genome-wide linkage analysis followed by dideoxy sequencing of the linked candidate gene LRAT was performed in a consanguineous Pakistani family with autosomal recessive retinitis pigmentosa. In silico prediction and minigene assays were used to investigate the effects of the presumptive splicing mutation.
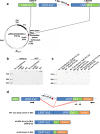
Results: ARRP in this family was linked to chromosome 4q31.21-q32.1 with a maximum LOD score of 5.40. A novel homozygous intronic mutation (NM_004744.4: c.541-15T>G) was detected in LRAT. In silico tools predicted that the AG-creating mutation would activate an intronic cryptic acceptor site, but cloning fragments of wild-type and mutant sequences of LRAT into Exontrap Cloning Vector pET01 and Expression Cloning Vector pCMV-(DYKD4K)-C showed that the primary effect of the sequence change was to weaken the nearby authentic acceptor site and cause exon skipping, with only a small fraction of transcripts utilizing the acceptor site producing the reference transcript.
Conclusions: The c.541-15T>G mutation in LRAT results in aberrant splicing and is therefore predicted to be causal for the early onset retinitis pigmentosa in this family. In addition, this work suggests that minigenes adapted to the specific gene and exon may need to be designed for variants in the first and last exon and intron to mimic the authentic splicing mechanism in vivo.
Keywords: Cryptic splice site; Exon splicing; LRAT; Linkage; Minigene assay; Retinitis pigmentosa; Splicing mutation.
Conflict of interest statement
Ethics approval and consent to participate
This study was approved by the Institutional Review Boards (IRB) of the National Centre of Excellence in Molecular Biology, Lahore, Pakistan, and the CNS IRB at the National Institutes of Health. Participating individuals or their guardians gave written informed consent to participate and to publish consistent with the tenets of the Declaration of Helsinki before the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Bird AC. Retinal photoreceptor dystrophies LI. Edward Jackson memorial lecture. AmJOphthalmol. 1995;119:543–562. - PubMed
-
- Bunker CH, Berson EL, Bromley WC, Hayes RP, Roderick TH. Prevalence of retinitis pigmentosa in Maine. AmJOphthalmol. 1984;97:357–365. - PubMed
-
- Rivolta C, Sharon D, DeAngelis MM, Dryja TP. Retinitis pigmentosa and allied diseases: numerous diseases, genes, and inheritance patterns. HumMolGenet. 2002;11:1219–1227. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

