Transcriptome Analysis of Adipose Tissue Indicates That the cAMP Signaling Pathway Affects the Feed Efficiency of Pigs
- PMID: 29973485
- PMCID: PMC6070815
- DOI: 10.3390/genes9070336
Transcriptome Analysis of Adipose Tissue Indicates That the cAMP Signaling Pathway Affects the Feed Efficiency of Pigs
Abstract
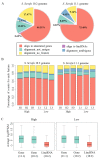
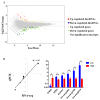
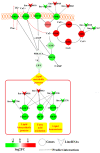
Feed efficiency (FE) is one of the main factors that determine the production costs in the pig industry. In this study, RNA Sequencing (RNA-seq) was applied to identify genes and long intergenic non-coding RNAs (lincRNAs) that are differentially expressed (DE) in the adipose tissues of Yorkshire pigs with extremely high and low FE. In total, 147 annotated genes and 18 lincRNAs were identified as DE between high- and low-FE pigs. Seventeen DE lincRNAs were significantly correlated with 112 DE annotated genes at the transcriptional level. Gene ontology (GO) analysis revealed that DE genes were significantly associated with cyclic adenosine monophosphate (cAMP) metabolic process and Ca2+ binding. cAMP, a second messenger has an important role in lipolysis, and its expression is influenced by Ca2+ levels. In high-FE pigs, nine DE genes with Ca2+ binding function, were down-regulated, whereas S100G, which encodes calbindin D9K that serve as a Ca2+ bumper, was up-regulated. Furthermore, ATP2B2, ATP1A4, and VIPR2, which participate in the cAMP signaling pathway, were down-regulated in the upstream of lipolysis pathways. In high-FE pigs, the key genes involved in the lipid biosynthetic process (ELOVL7 and B4GALT6), fatty acid oxidation (ABCD2 and NR4A3), and lipid homeostasis (C1QTNF3 and ABCB4) were down-regulated. These results suggested that cAMP was involved in the regulation on FE of pigs by affecting lipid metabolism in adipose tissues.
Keywords: adipose; cAMP; feed efficiency; lipid metabolism; pig.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Transcriptome Analysis Reveals that Vitamin A Metabolism in the Liver Affects Feed Efficiency in Pigs.G3 (Bethesda). 2016 Nov 8;6(11):3615-3624. doi: 10.1534/g3.116.032839. G3 (Bethesda). 2016. PMID: 27633790 Free PMC article.
-
Transcriptome Analysis Reveals Long Intergenic Non-Coding RNAs Contributed to Intramuscular Fat Content Differences between Yorkshire and Wei Pigs.Int J Mol Sci. 2020 Mar 3;21(5):1732. doi: 10.3390/ijms21051732. Int J Mol Sci. 2020. PMID: 32138348 Free PMC article.
-
Transcriptome analysis of adipose tissue from pigs divergent in feed efficiency reveals alteration in gene networks related to adipose growth, lipid metabolism, extracellular matrix, and immune response.Mol Genet Genomics. 2019 Apr;294(2):395-408. doi: 10.1007/s00438-018-1515-5. Epub 2018 Nov 27. Mol Genet Genomics. 2019. PMID: 30483895
-
Neuronal Signal Transduction-Involved Genes in Pig Hypothalamus Affect Feed Efficiency as Revealed by Transcriptome Analysis.Biomed Res Int. 2018 Dec 26;2018:5862571. doi: 10.1155/2018/5862571. eCollection 2018. Biomed Res Int. 2018. PMID: 30687750 Free PMC article.
-
Role of the cAMP Pathway in Glucose and Lipid Metabolism.Handb Exp Pharmacol. 2016;233:29-49. doi: 10.1007/164_2015_32. Handb Exp Pharmacol. 2016. PMID: 26721678 Review.
Cited by
-
Effects of Cyclic Adenosine Monophosphate Nanoliposomes on Growth Performance, Gut Development and Microbiota of Broilers.Animals (Basel). 2025 Jun 23;15(13):1852. doi: 10.3390/ani15131852. Animals (Basel). 2025. PMID: 40646751 Free PMC article.
-
The Effects of Naringenin on miRNA-mRNA Profiles in HepaRG Cells.Int J Mol Sci. 2021 Feb 25;22(5):2292. doi: 10.3390/ijms22052292. Int J Mol Sci. 2021. PMID: 33669020 Free PMC article.
-
Identification of Differentially Expressed miRNAs in Porcine Adipose Tissues and Evaluation of Their Effects on Feed Efficiency.Genes (Basel). 2022 Dec 19;13(12):2406. doi: 10.3390/genes13122406. Genes (Basel). 2022. PMID: 36553673 Free PMC article.
-
Investigation of muscle transcriptomes using gradient boosting machine learning identifies molecular predictors of feed efficiency in growing pigs.BMC Genomics. 2019 Aug 17;20(1):659. doi: 10.1186/s12864-019-6010-9. BMC Genomics. 2019. PMID: 31419934 Free PMC article.
-
DNA Imprinting and Differentially Expressed Genes in Longissimus thoracis Muscle of Bos indicus Submitted to Early Weaning Management.Epigenomes. 2024 Dec 4;8(4):45. doi: 10.3390/epigenomes8040045. Epigenomes. 2024. PMID: 39727807 Free PMC article.
References
-
- Lkhagvadorj S., Qu L., Cai W., Couture O.P., Barb C.R., Hausman G.J., Nettleton D., Anderson L.L., Dekkers J.C., Tuggle C.K. Gene expression profiling of the short-term adaptive response to acute caloric restriction in liver and adipose tissues of pigs differing in feed efficiency. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010;298:R494–R507. doi: 10.1152/ajpregu.00632.2009. - DOI - PubMed
-
- Louveau I., Vincent A., Tacher S., Gilbert H., Gondret F. Increased expressions of genes and proteins involved in mitochondrial oxidation and antioxidant pathway in adipose tissue of pigs selected for a low residual feed intake. J. Anim. Sci. 2016;94:5042–5054. doi: 10.2527/jas.2016-0619. - DOI - PubMed
-
- Gondret F., Vincent A., Houee-Bigot M., Siegel A., Lagarrigue S., Causeur D., Gilbert H., Louveau I. A transcriptome multi-tissue analysis identifies biological pathways and genes associated with variations in feed efficiency of growing pigs. BMC Genom. 2017;18:244. doi: 10.1186/s12864-017-3639-0. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

