Dissecting Distinct Roles of NEDDylation E1 Ligase Heterodimer APPBP1 and UBA3 Reveals Potential Evolution Process for Activation of Ubiquitin-related Pathways
- PMID: 29973603
- PMCID: PMC6031683
- DOI: 10.1038/s41598-018-28214-2
Dissecting Distinct Roles of NEDDylation E1 Ligase Heterodimer APPBP1 and UBA3 Reveals Potential Evolution Process for Activation of Ubiquitin-related Pathways
Abstract
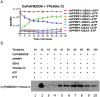
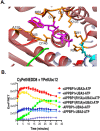
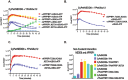
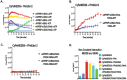
Despite the similar enzyme cascade in the Ubiquitin and Ubiquitin-like peptide(Ubl) conjugation, the involvement of single or heterodimer E1 activating enzyme has been a mystery. Here, by using a quantitative Förster Resonance Energy Transfer (FRET) technology, aided with Analysis of Electrostatic Similarities Of Proteins (AESOP) computational framework, we elucidate in detail the functional properties of each subunit of the E1 heterodimer activating-enzyme for NEDD8, UBA3 and APPBP1. In contrast to SUMO activation, which requires both subunits of its E1 heterodimer AOS1-Uba2 for its activation, NEDD8 activation requires only one of two E1 subunits, UBA3. The other subunit, APPBP1, only contributes by accelerating the activation reaction rate. This discovery implies that APPBP1 functions mainly as a scaffold protein to enhance molecular interactions and facilitate catalytic reaction. These findings for the first time reveal critical new mechanisms and a potential evolutionary pathway for Ubl activations. Furthermore, this quantitative FRET approach can be used for other general biochemical pathway analysis in a dynamic mode.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Structural dissection of a gating mechanism preventing misactivation of ubiquitin by NEDD8's E1.Biochemistry. 2008 Aug 26;47(34):8961-9. doi: 10.1021/bi800604c. Epub 2008 Jul 25. Biochemistry. 2008. PMID: 18652489 Free PMC article.
-
The structure of the APPBP1-UBA3-NEDD8-ATP complex reveals the basis for selective ubiquitin-like protein activation by an E1.Mol Cell. 2003 Dec;12(6):1427-37. doi: 10.1016/s1097-2765(03)00452-0. Mol Cell. 2003. PMID: 14690597
-
Conservation in the mechanism of Nedd8 activation by the human AppBp1-Uba3 heterodimer.J Biol Chem. 2003 Jul 18;278(29):26823-30. doi: 10.1074/jbc.M303177200. Epub 2003 May 10. J Biol Chem. 2003. PMID: 12740388
-
Ubiquitin-like Protein Conjugation: Structures, Chemistry, and Mechanism.Chem Rev. 2018 Feb 14;118(3):889-918. doi: 10.1021/acs.chemrev.6b00737. Epub 2017 Feb 24. Chem Rev. 2018. PMID: 28234446 Free PMC article. Review.
-
Targeting DCN1-UBC12 Protein-Protein Interaction for Regulation of Neddylation Pathway.Adv Exp Med Biol. 2020;1217:349-362. doi: 10.1007/978-981-15-1025-0_20. Adv Exp Med Biol. 2020. PMID: 31898237 Review.
Cited by
-
The Protein Response of Salt-Tolerant Zygosaccharomyces rouxii to High-Temperature Stress during the Lag Phase.J Fungi (Basel). 2024 Jan 5;10(1):48. doi: 10.3390/jof10010048. J Fungi (Basel). 2024. PMID: 38248957 Free PMC article.
-
Targeting neddylation E2s: a novel therapeutic strategy in cancer.J Hematol Oncol. 2021 Apr 7;14(1):57. doi: 10.1186/s13045-021-01070-w. J Hematol Oncol. 2021. PMID: 33827629 Free PMC article. Review.
-
FOXO1-regulated lncRNA CYP1B1-AS1 suppresses breast cancer cell proliferation by inhibiting neddylation.Breast Cancer Res Treat. 2023 Nov;202(2):397-408. doi: 10.1007/s10549-023-07090-z. Epub 2023 Aug 28. Breast Cancer Res Treat. 2023. PMID: 37640964 Free PMC article.
-
The NEDD8-activating enzyme E1 UBA3 orchestrates the immunosuppressive microenvironment in lung adenocarcinoma via the NF-кB pathway.Med Oncol. 2023 Sep 1;40(10):286. doi: 10.1007/s12032-023-02162-y. Med Oncol. 2023. PMID: 37656220 Free PMC article.
-
Bi-allelic variants in NAE1 cause intellectual disability, ischiopubic hypoplasia, stress-mediated lymphopenia and neurodegeneration.Am J Hum Genet. 2023 Jan 5;110(1):146-160. doi: 10.1016/j.ajhg.2022.12.003. Am J Hum Genet. 2023. PMID: 36608681 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

