Phosphorylation Modulates the Subcellular Localization of SOX11
- PMID: 29973868
- PMCID: PMC6020773
- DOI: 10.3389/fnmol.2018.00211
Phosphorylation Modulates the Subcellular Localization of SOX11
Abstract
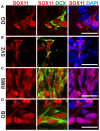
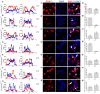
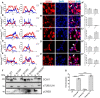
SOX11 is a key Transcription Factor (TF) in the regulation of embryonic and adult neurogenesis, whose mutation has recently been linked to an intellectual disability syndrome in humans. SOX11's transient activity during neurogenesis is critical to ensure the precise execution of the neurogenic program. Here, we report that SOX11 displays differential subcellular localizations during the course of neurogenesis. Western-Blot analysis of embryonic mouse brain lysates indicated that SOX11 is post-translationally modified by phosphorylation. Using Mass Spectrometry, we found 10 serine residues in the SOX11 protein that are putatively phosphorylated. Systematic analysis of phospho-mutant SOX11 resulted in the identification of the S30 residue, whose phosphorylation promotes nuclear over cytoplasmic localization of SOX11. Collectively, these findings uncover phosphorylation as a novel layer of regulation of the intellectual disability gene Sox11.
Keywords: SOX11; cancer; intellectual disability; neurogenesis; subcellular localization; transcription factor phosphorylation.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

