Functional Genetic Diversity and Culturability of Petroleum-Degrading Bacteria Isolated From Oil-Contaminated Soils
- PMID: 29973925
- PMCID: PMC6019457
- DOI: 10.3389/fmicb.2018.01332
Functional Genetic Diversity and Culturability of Petroleum-Degrading Bacteria Isolated From Oil-Contaminated Soils
Abstract
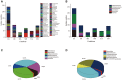
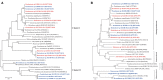
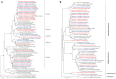
In this study, we compared the culturability of aerobic bacteria isolated from long-term oil-contaminated soils via enrichment and direct-plating methods; bacteria were cultured at 30°C or ambient temperatures. Two soil samples were collected from two sites in the Shengli oilfield located in Dongying, China. One sample (S0) was close to the outlet of an oil-production water treatment plant, and the other sample (S1) was located 500 m downstream of the outlet. In total, 595 bacterial isolates belonging to 56 genera were isolated, distributed in Actinobacteria, Firmicutes, Bacterioidetes, and Proteobacteria. It was interesting that Actinobacteria and Firmicutes were not detected from the 16S rRNA gene clone library. The results suggested the activation of rare species during culture. Using the enrichment method, 239 isolates (31 genera) and 96 (22 genera) isolates were obtained at ambient temperatures and 30°C, respectively, from S0 soil. Using the direct-plating method, 97 isolates (15 genera) and 163 isolates (20 genera) were obtained at ambient temperatures and 30°C, respectively, from two soils. Of the 595 isolates, 244 isolates (41.7% of total isolates) could degrade n-hexadecane. A greater number of alkane-degraders was isolated at ambient temperatures using the enrichment method, suggesting that this method could significantly improve bacterial culturability. Interestingly, the proportion of alkane degrading isolates was lower in the isolates obtained using enrichment method than that obtained using direct-plating methods. Considering the greater species diversity of isolates obtained via the enrichment method, this technique could be used to increase the diversity of the microbial consortia. Furthermore, phenol hydroxylase genes (pheN), medium-chain alkane monooxygenases genes (alkB and CYP153A), and long-chain alkane monooxygenase gene (almA) were detected in 60 isolates (11 genotypes), 91 isolates (27 genotypes) and 93 isolates (24 genotypes), and 34 isolates (14 genotypes), respectively. This study could provide new insights into microbial resources from oil fields or other environments, and this information will be beneficial for bioremediation of petroleum contamination and for other industrial applications.
Keywords: ambient temperature; bacteria; bioremediation; culturability; petroleum degradation.
Figures


Similar articles
-
Diversity of crude oil-degrading bacteria and alkane hydroxylase (alkB) genes from the Qinghai-Tibet Plateau.Environ Monit Assess. 2017 Mar;189(3):116. doi: 10.1007/s10661-017-5798-5. Epub 2017 Feb 20. Environ Monit Assess. 2017. PMID: 28220441
-
Gene diversity of CYP153A and AlkB alkane hydroxylases in oil-degrading bacteria isolated from the Atlantic Ocean.Environ Microbiol. 2010 May;12(5):1230-42. doi: 10.1111/j.1462-2920.2010.02165.x. Epub 2010 Feb 11. Environ Microbiol. 2010. PMID: 20148932
-
Diversity of alkane hydroxylase genes on the rhizoplane of grasses planted in petroleum-contaminated soils.Springerplus. 2015 Sep 18;4:526. doi: 10.1186/s40064-015-1312-0. eCollection 2015. Springerplus. 2015. PMID: 26405645 Free PMC article.
-
Microbial Degradation of Rubber: Actinobacteria.Polymers (Basel). 2021 Jun 17;13(12):1989. doi: 10.3390/polym13121989. Polymers (Basel). 2021. PMID: 34204568 Free PMC article. Review.
-
Survival and Energy Producing Strategies of Alkane Degraders Under Extreme Conditions and Their Biotechnological Potential.Front Microbiol. 2018 May 25;9:1081. doi: 10.3389/fmicb.2018.01081. eCollection 2018. Front Microbiol. 2018. PMID: 29910779 Free PMC article. Review.
Cited by
-
Characterization of the biosurfactant production and enzymatic potential of bacteria isolated from an oil-contaminated saline soil.Int Microbiol. 2023 Aug;26(3):529-542. doi: 10.1007/s10123-022-00318-w. Epub 2023 Jan 21. Int Microbiol. 2023. PMID: 36680696
-
Polyethylene Biodegradation by an Artificial Bacterial Consortium: Rhodococcus as a Competitive Plastisphere Species.Microbes Environ. 2024;39(3):ME24031. doi: 10.1264/jsme2.ME24031. Microbes Environ. 2024. PMID: 39085141 Free PMC article.
-
Genome Sequence of Trichoderma lixii MUT3171, A Promising Strain for Mycoremediation of PAH-Contaminated Sites.Microorganisms. 2020 Aug 20;8(9):1258. doi: 10.3390/microorganisms8091258. Microorganisms. 2020. PMID: 32825267 Free PMC article.
-
Microbial Metabolic Potential of Phenol Degradation in Wastewater Treatment Plant of Crude Oil Refinery: Analysis of Metagenomes and Characterization of Isolates.Microorganisms. 2020 Apr 30;8(5):652. doi: 10.3390/microorganisms8050652. Microorganisms. 2020. PMID: 32365784 Free PMC article.
-
Integrated Comparative Genomic Analysis and Phenotypic Profiling of Pseudomonas aeruginosa Isolates From Crude Oil.Front Microbiol. 2020 Mar 31;11:519. doi: 10.3389/fmicb.2020.00519. eCollection 2020. Front Microbiol. 2020. PMID: 32300337 Free PMC article.
References
-
- Barbe V., Vallenet D., Fonknechten N., Kreimeyer A., Oztas S., Labarre L., et al. (2004). Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32, 5766–5779. 10.1093/nar/gkh910 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

