Activation of the Immune-Metabolic Receptor GPR84 Enhances Inflammation and Phagocytosis in Macrophages
- PMID: 29973940
- PMCID: PMC6019444
- DOI: 10.3389/fimmu.2018.01419
Activation of the Immune-Metabolic Receptor GPR84 Enhances Inflammation and Phagocytosis in Macrophages
Abstract
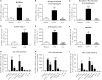
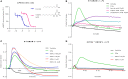
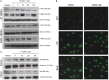
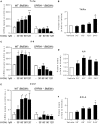
GPR84 is a member of the metabolic G protein-coupled receptor family, and its expression has been described predominantly in immune cells. GPR84 activation is involved in the inflammatory response, but the mechanisms by which it modulates inflammation have been incompletely described. In this study, we investigated GPR84 expression, activation, and function in macrophages to establish the role of the receptor during the inflammatory response. We observed that GPR84 expression in murine tissues is increased by endotoxemia, hyperglycemia, and hypercholesterolemia. Ex vivo studies revealed that GPR84 mRNA expression is increased by LPS and other pro-inflammatory molecules in different murine and human macrophage populations. Likewise, high glucose concentrations and the presence of oxidized LDL increased GPR84 expression in macrophages. Activation of the GPR84 receptor with a selective agonist, 6-(octylamino) pyrimidine-2,4(1H,3H)-dione (6-n-octylaminouracil, 6-OAU), enhanced the expression of phosphorylated Akt, p-ERK, and p65 nuclear translocation under inflammatory conditions and elevated the expression levels of the inflammatory mediators TNFα, IL-6, IL-12B, CCL2, CCL5, and CXCL1. In addition, GPR84 activation triggered increased bacterial adhesion and phagocytosis in macrophages. The enhanced inflammatory response mediated by 6-OAU was not observed in GPR84-/- cells nor in macrophages treated with a selective GPR84 antagonist. Collectively, our results reveal that GPR84 functions as an enhancer of inflammatory signaling in macrophages once inflammation is established. Therefore, molecules that antagonize the GPR84 receptor may be potential therapeutic tools in inflammatory and metabolic diseases.
Keywords: GPR84; immunometabolism; inflammation; macrophages; metabolic G protein-coupled receptors.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

