A current view on long noncoding RNAs in yeast and filamentous fungi
- PMID: 29974182
- PMCID: PMC6097775
- DOI: 10.1007/s00253-018-9187-y
A current view on long noncoding RNAs in yeast and filamentous fungi
Abstract
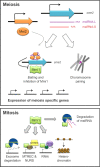
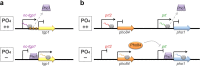
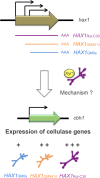
Long noncoding RNAs (lncRNAs) are crucial players in epigenetic regulation. They were initially discovered in human, yet they emerged as common factors involved in a number of central cellular processes in several eukaryotes. For example, in the past decade, research on lncRNAs in yeast has steadily increased. Several examples of lncRNAs were described in Saccharomyces cerevisiae and Schizosaccharomyces pombe. Also, screenings for lncRNAs in ascomycetes were performed and, just recently, the first full characterization of a lncRNA was performed in the filamentous fungus Trichoderma reesei. In this review, we provide a broad overview about currently known fugal lncRNAs. We make an attempt to categorize them according to their functional context, regulatory strategies or special properties. Moreover, the potential of lncRNAs as a biotechnological tool is discussed.
Keywords: Long noncoding RNA; Saccharomyces cerevisiae; Schizosaccharomyces pombe; Trichoderma reesei; Yeast; lncRNA.
Conflict of interest statement
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Figures
Similar articles
-
The long non-coding RNA world in yeasts.Biochim Biophys Acta. 2016 Jan;1859(1):147-54. doi: 10.1016/j.bbagrm.2015.08.003. Epub 2015 Aug 8. Biochim Biophys Acta. 2016. PMID: 26265144 Review.
-
Long Noncoding RNAs in the Yeast S. cerevisiae.Adv Exp Med Biol. 2017;1008:119-132. doi: 10.1007/978-981-10-5203-3_4. Adv Exp Med Biol. 2017. PMID: 28815538 Review.
-
Long noncoding RNA-based chromatin control of germ cell differentiation: a yeast perspective.Chromosome Res. 2013 Dec;21(6-7):653-63. doi: 10.1007/s10577-013-9393-5. Chromosome Res. 2013. PMID: 24249577 Free PMC article. Review.
-
cDNA encoding protein O-mannosyltransferase from the filamentous fungus Trichoderma reesei; functional equivalence to Saccharomyces cerevisiae PMT2.Curr Genet. 2003 Apr;43(1):11-6. doi: 10.1007/s00294-003-0368-5. Epub 2003 Feb 13. Curr Genet. 2003. PMID: 12684840
-
Dolichol phosphate mannose synthase from the filamentous fungus Trichoderma reesei belongs to the human and Schizosaccharomyces pombe class of the enzyme.Glycobiology. 2000 Oct;10(10):983-91. doi: 10.1093/glycob/10.10.983. Glycobiology. 2000. PMID: 11030744
Cited by
-
Differential expression patterns of long noncoding RNAs in a pleiomorphic diatom and relation to hyposalinity.Sci Rep. 2023 Feb 10;13(1):2440. doi: 10.1038/s41598-023-29489-w. Sci Rep. 2023. PMID: 36765079 Free PMC article.
-
Advances in understanding the evolution of fungal genome architecture.F1000Res. 2020 Jul 27;9:F1000 Faculty Rev-776. doi: 10.12688/f1000research.25424.1. eCollection 2020. F1000Res. 2020. PMID: 32765832 Free PMC article. Review.
-
Exosomal Long Non-coding RNAs: Emerging Players in the Tumor Microenvironment.Mol Ther Nucleic Acids. 2020 Oct 4;23:1371-1383. doi: 10.1016/j.omtn.2020.09.039. eCollection 2021 Mar 5. Mol Ther Nucleic Acids. 2020. PMID: 33738133 Free PMC article. Review.
-
Identification and characterization of pineapple leaf lncRNAs in crassulacean acid metabolism (CAM) photosynthesis pathway.Sci Rep. 2019 Apr 30;9(1):6658. doi: 10.1038/s41598-019-43088-8. Sci Rep. 2019. PMID: 31040312 Free PMC article.
-
The long non-coding RNA landscape of Candida yeast pathogens.Nat Commun. 2021 Dec 16;12(1):7317. doi: 10.1038/s41467-021-27635-4. Nat Commun. 2021. PMID: 34916523 Free PMC article.
References
-
- Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, Fink GR. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol Cell. 2012;45(4):470–482. doi: 10.1016/j.molcel.2011.11.029. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

