LTR-retrotransposon transcriptome modulation in response to endotoxin-induced stress in PBMCs
- PMID: 29976163
- PMCID: PMC6034278
- DOI: 10.1186/s12864-018-4901-9
LTR-retrotransposon transcriptome modulation in response to endotoxin-induced stress in PBMCs
Abstract
Background: Human Endogenous Retroviruses (HERVs) and Mammalian apparent LTR-retrotransposons (MaLRs) represent the 8% of our genome and are distributed among our 46 chromosomes. These LTR-retrotransposons are thought to be essentially silent except in cancer, autoimmunity and placental development. Their Long Terminal Repeats (LTRs) constitute putative promoter or polyA regulatory sequences. In this study, we used a recently described high-density microarray which can be used to study HERV/MaLR transcriptome including 353,994 HERV/MaLR loci and 1559 immunity-related genes.
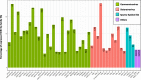
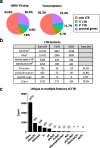
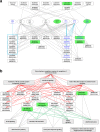
Results: We described, for the first time, the HERV transcriptome in peripheral blood mononuclear cells (PBMCs) using a cellular model mimicking inflammatory response and monocyte anergy observed after septic shock. About 5.6% of the HERV/MaLR repertoire is transcribed in PBMCs. Roughly one-tenth [5.7-13.1%] of LTRs exhibit a putative constitutive promoter or polyA function while one-quarter [19.5-27.6%] may shift from silent to active. Evidence was given that some HERVs/MaLRs and genes may share similar regulation control under lipopolysaccharide (LPS) stimulation conditions. Stimulus-dependent response confirms that HERV expression is tightly regulated in PBMCs. Altogether, these observations make it possible to integrate 62 HERVs/MaLRs and 26 genes in 11 canonical pathways and suggest a link between HERV expression and immune response. The transcriptional modulation of HERVs located close to genes such as OAS2/3 and IFI44/IFI44L or at a great distance from genes was discussed.
Conclusion: This microarray-based approach revealed the expression of about 47,466 distinct HERV loci and identified 951 putative promoter LTRs and 744 putative polyA LTRs in PBMCs. HERV/MaLR expression was shown to be tightly modulated under several stimuli including high-dose and low-dose LPS and Interferon-γ (IFN-γ). HERV incorporation at the crossroads of immune response pathways paves the way for further functional studies and analyses of the HERV transcriptome in altered immune responses in vivo such as in sepsis.
Keywords: Endotoxin tolerance; HERV transcriptome; LPS; PBMCs; Sepsis; Signalling pathways.
Conflict of interest statement
Ethics approval and consent to participate
Citrated pouches or heparinised tubes blood were obtained from EFS (Etablissement Français du Sang) and used immediately. According to EFS standardised procedures for blood donation and to provisions of the articles R.1243–49 and following ones of the French Public Health Code, a written non-opposition to the use of his donation for research purposes was obtained from healthy volunteers. The blood donors’ personal data were anonymised before blood transfer to our research lab. We obtained the favourable notice of the Local Ethical Committee (Comité de Protection des Personnes Sud-Est II, Bâtiment Pinel, 59 Boulevard Pinel, 69,500 Bron) and the acceptance of the Ministère de la Recherche (declaration DC-2008-64) for handling and conservation of these samples.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Perot P, Bolze P-A, Mallet F. From viruses to genes: syncytins. In: Witzany G, editor. Viruses, Essential Agents of Life. Netherlands: Springer; 2012. p. 325–61.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

