Metagenomic analysis of basal ice from an Alaskan glacier
- PMID: 29976249
- PMCID: PMC6034282
- DOI: 10.1186/s40168-018-0505-5
Metagenomic analysis of basal ice from an Alaskan glacier
Abstract
Background: Glaciers cover ~ 10% of land but are among the least explored environments on Earth. The basal portion of glaciers often harbors unique aquatic microbial ecosystems in the absence of sunlight, and knowledge on the microbial community structures and their metabolic potential is very limited. Here, we provide insights into the microbial lifestyle present at the base of the Matanuska Glacier, Alaska.
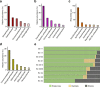
Results: DNA and RNA were extracted from samples of the Matanuska Glacier basal ice. Using Illumina MiSeq and HiSeq sequencing, we investigated the microbial diversity with the metagenomic shotgun reads and 16S ribosomal RNA data. We further assembled 9 partial and draft bacterial genomes from the metagenomic assembly, and identified key metabolic pathways such as sulfur oxidation and nitrification. Collectively, our analyses suggest a prevalence of lithotrophic and heterotrophic metabolisms in the subglacial microbiome.
Conclusion: Our results present the first metagenomic assembly and bacterial draft genomes for a subglacial environment. These results extend our understanding of the chemical and biological processes in subglacial environments critically influenced by global climate change.
Keywords: Basal ice layer; Glacier; Metagenomics; Microbiome.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
References
-
- Horgan HJ, Alley RB, Christianson K, Jacobel RW, Anandakrishnan S, Muto A, Beem LH, Siegfried MR. Estuaries beneath ice sheets. Geology. 2013;41(11):1159–1162. doi: 10.1130/G34654.1. - DOI
-
- Wadham JL, Tranter M, Skidmore M, Hodson AJ, Priscu J, Lyons WB, Sharp M, Wynn P, Jackson M. Biogeochemical weathering under ice: size matters. Glob Biogeochem Cycles. 2010;24(3). 10.1029/2009GB003688.
-
- Skidmore M. Microbial communities in Antarctic subglacial aquatic environments. In: Antarctic subglacial aquatic environments. Washington DC: American Geophysical Union; 2011. p. 61–81.
-
- Achberger AM, Michaud AB, Vick-Majors TJ, Christner BC, Skidmore ML, Priscu JC, Tranter M. Microbiology of subglacial environments. In: Psychrophiles: from biodiversity to biotechnology. Cham: Springer; 2017. p. 83–110.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

