Replication Stress Drives Constitutive Activation of the DNA Damage Response and Radioresistance in Glioblastoma Stem-like Cells
- PMID: 29976574
- PMCID: PMC6128404
- DOI: 10.1158/0008-5472.CAN-18-0569
Replication Stress Drives Constitutive Activation of the DNA Damage Response and Radioresistance in Glioblastoma Stem-like Cells
Abstract
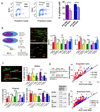
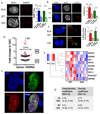
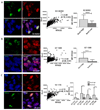
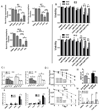
Glioblastoma (GBM) is a lethal primary brain tumor characterized by treatment resistance and inevitable tumor recurrence, both of which are driven by a subpopulation of GBM cancer stem-like cells (GSC) with tumorigenic and self-renewal properties. Despite having broad implications for understanding GSC phenotype, the determinants of upregulated DNA-damage response (DDR) and subsequent radiation resistance in GSC are unknown and represent a significant barrier to developing effective GBM treatments. In this study, we show that constitutive DDR activation and radiation resistance are driven by high levels of DNA replication stress (RS). CD133+ GSC exhibited reduced DNA replication velocity and a higher frequency of stalled replication forks than CD133- non-GSC in vitro; immunofluorescence studies confirmed these observations in a panel of orthotopic xenografts and human GBM specimens. Exposure of non-GSC to low-level exogenous RS generated radiation resistance in vitro, confirming RS as a novel determinant of radiation resistance in tumor cells. GSC exhibited DNA double-strand breaks, which colocalized with "replication factories" and RNA: DNA hybrids. GSC also demonstrated increased expression of long neural genes (>1 Mbp) containing common fragile sites, supporting the hypothesis that replication/transcription collisions are the likely cause of RS in GSC. Targeting RS by combined inhibition of ATR and PARP (CAiPi) provided GSC-specific cytotoxicity and complete abrogation of GSC radiation resistance in vitro These data identify RS as a cancer stem cell-specific target with significant clinical potential.Significance: These findings shed new light on cancer stem cell biology and reveal novel therapeutics with the potential to improve clinical outcomes by overcoming inherent radioresistance in GBM. Cancer Res; 78(17); 5060-71. ©2018 AACR.
©2018 American Association for Cancer Research.
Conflict of interest statement
Figures

Comment in
-
Replication Stress: An Achilles' Heel of Glioma Cancer Stem-like Cells.Cancer Res. 2018 Dec 15;78(24):6713-6716. doi: 10.1158/0008-5472.CAN-18-2439. Epub 2018 Nov 29. Cancer Res. 2018. PMID: 30498082 Free PMC article.
References
-
- Ajaz M, Jefferies S, Brazil L, Watts C, Chalmers A. Current and investigational drug strategies for glioblastoma. Clin Oncol (R Coll Radiol) 2014;26(7):419–30. - PubMed
-
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–96. - PubMed
-
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–60. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

