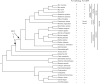
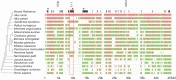
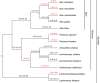
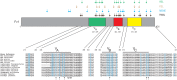
Ancient Evolutionary Origin and Positive Selection of the Retroviral Restriction Factor Fv1 in Muroid Rodents
- PMID: 29976659
- PMCID: PMC6146698
- DOI: 10.1128/JVI.00850-18
Ancient Evolutionary Origin and Positive Selection of the Retroviral Restriction Factor Fv1 in Muroid Rodents
Abstract
The laboratory mouse Fv1 gene encodes a retroviral restriction factor that mediates resistance to murine leukemia viruses (MLVs). Sequence similarity between Fv1 and the gag protein of the murine endogenous retrovirus L (MuERV-L) family of ERVs suggests that Fv1 was coopted from an ancient provirus. Previous evolutionary studies found Fv1 orthologs only in the genus Mus Here, we describe identification of orthologous Fv1 sequences in several species belonging to multiple families of rodents outside the genus Mus We show that these Fv1 orthologs are in the same region of conserved synteny, between the genes Miip and Mfn2, suggesting a minimum insertion time of 45 million years for the ancient progenitor of Fv1 Our analysis also revealed that Fv1 was not detectable or heavily mutated in some lineages in the superfamily Muroidea, while, in concert with previous findings in the genus Mus, we found strong evidence of positive selection of Fv1 in the African clade in the subfamily Muridae Residues identified as evolving under positive selection include those that have been previously found to be important for restriction of multiple retroviral lineages. Taken together, these findings suggest that the evolutionary origin of Fv1 substantially predates Mus evolution, that the rodent Fv1 has been shaped by lineage-specific differential selection pressures, and that Fv1 has long been evolving under positive selection in the rodent family Muridae, supporting a defensive role that significantly antedates exposure to MLVs.IMPORTANCE Retroviruses have adapted to living in concert with their hosts throughout vertebrate evolution. Over the years, the study of these relationships revealed the presence of host proteins called restriction factors that inhibit retroviral replication in host cells. The first of these restriction factors to be identified, encoded by the Fv1 gene found in mice, was thought to have originated in the genus Mus In this study, we utilized genome database searches and DNA sequencing to identify Fv1 copies in multiple rodent lineages. Our findings suggest a minimum time of insertion into the genome of rodents of 45 million years for the ancestral progenitor of Fv1 While Fv1 is not detectable in some lineages, we also identified full-length orthologs showing signatures of a molecular "arms race" in a family of rodent species indigenous to Africa. This finding suggests that Fv1 in these species has been coevolving with unidentified retroviruses for millions of years.
Keywords: Fv1 restriction factor; gammaretrovirus restriction; mouse leukemia viruses; rodent evolution.
Copyright © 2018 American Society for Microbiology.
Figures

Similar articles
-
Origin, antiviral function and evidence for positive selection of the gammaretrovirus restriction gene Fv1 in the genus Mus.Proc Natl Acad Sci U S A. 2009 Mar 3;106(9):3259-63. doi: 10.1073/pnas.0900181106. Epub 2009 Feb 12. Proc Natl Acad Sci U S A. 2009. PMID: 19221034 Free PMC article.
-
Evolutionary journey of the retroviral restriction gene Fv1.Proc Natl Acad Sci U S A. 2018 Oct 2;115(40):10130-10135. doi: 10.1073/pnas.1808516115. Epub 2018 Sep 17. Proc Natl Acad Sci U S A. 2018. PMID: 30224488 Free PMC article.
-
Patterns of Coevolutionary Adaptations across Time and Space in Mouse Gammaretroviruses and Three Restrictive Host Factors.Viruses. 2021 Sep 18;13(9):1864. doi: 10.3390/v13091864. Viruses. 2021. PMID: 34578445 Free PMC article.
-
Fv1, the mouse retrovirus resistance gene.Rev Sci Tech. 1998 Apr;17(1):269-77. doi: 10.20506/rst.17.1.1080. Rev Sci Tech. 1998. PMID: 9638816 Review.
-
Restriction factors: a defense against retroviral infection.Trends Microbiol. 2003 Jun;11(6):286-91. doi: 10.1016/s0966-842x(03)00123-9. Trends Microbiol. 2003. PMID: 12823946 Review.
Cited by
-
Heterozygous and generalist MxA super-restrictors overcome breadth-specificity tradeoffs in antiviral restriction.bioRxiv [Preprint]. 2024 Oct 10:2024.10.10.617484. doi: 10.1101/2024.10.10.617484. bioRxiv. 2024. Update in: Sci Adv. 2025 May 2;11(18):eadu0062. doi: 10.1126/sciadv.adu0062. PMID: 39416221 Free PMC article. Updated. Preprint.
-
Evolutionary conservation of an ancient retroviral gagpol gene in Artiodactyla.J Virol. 2023 Sep 28;97(9):e0053523. doi: 10.1128/jvi.00535-23. Epub 2023 Sep 5. J Virol. 2023. PMID: 37668369 Free PMC article.
-
In-silico structural analysis of Heterocephalus glaber amyloid beta: an anti-Alzheimer's peptide.Mol Biol Res Commun. 2024;13(1):29-42. doi: 10.22099/mbrc.2023.48223.1862. Mol Biol Res Commun. 2024. PMID: 38164365 Free PMC article.
-
Transcriptional Regulation of Endogenous Retroviruses and Their Misregulation in Human Diseases.Int J Mol Sci. 2022 Sep 4;23(17):10112. doi: 10.3390/ijms231710112. Int J Mol Sci. 2022. PMID: 36077510 Free PMC article. Review.
-
Crosstalk between the Intestinal Virome and Other Components of the Microbiota, and Its Effect on Intestinal Mucosal Response and Diseases.J Immunol Res. 2022 Sep 27;2022:7883945. doi: 10.1155/2022/7883945. eCollection 2022. J Immunol Res. 2022. PMID: 36203793 Free PMC article. Review.
References
-
- Benit L, De Parseval N, Casella JF, Callebaut I, Cordonnier A, Heidmann T. 1997. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J Virol 71:5652–5657. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

