Interaction via the N terminus of the type IV secretion system (T4SS) protein VirB6 with VirB10 is required for VirB2 and VirB5 incorporation into T-pili and for T4SS function
- PMID: 29976757
- PMCID: PMC6120205
- DOI: 10.1074/jbc.RA118.002751
Interaction via the N terminus of the type IV secretion system (T4SS) protein VirB6 with VirB10 is required for VirB2 and VirB5 incorporation into T-pili and for T4SS function
Abstract
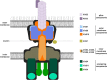
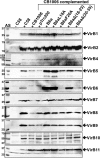
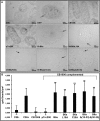
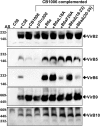
Many bacterial pathogens employ multicomponent protein complexes such as type IV secretion systems (T4SSs) to transfer virulence factors into host cells. Here we studied the interaction between two essential T4SS components: the very hydrophobic inner membrane protein VirB6, which may be a component of the translocation channel, and VirB10, which links the inner and outer bacterial membranes. To map the interaction site between these two T4SS components, we conducted alanine scanning and deleted six-amino acid stretches from the N-terminal periplasmic domain of VirB6 from Brucella suis Using the bacterial two-hybrid system to analyze the effects of these alterations on the VirB6-VirB10 interaction, we identified the amino acid regions 16-21 and 28-33 and Leu-18 in VirB6 as being required for this interaction. SDS-PAGE coupled with Western blotting of cell lysates and native PAGE of detergent-extracted membrane proteins revealed that the corresponding VirB6 residues in Agrobacterium tumefaciens (Phe-20 and amino acids 18-23 and 30-35) modulate the stability of both VirB6 and VirB5. However, the results from immuno-EM and super-resolution microscopy suggested that these regions and residues are not required for membrane association or for polar localization of VirB6. The six-amino acid deletions in the N terminus of VirB6 abolished pilus formation and virulence of A. tumefaciens, and the corresponding deletions in the VirB6 homolog TraD from the plasmid pKM101-T4SS abrogated plasmid transfer. Our results indicate that specific residues of the VirB6 N-terminal domain are required for VirB6 stabilization, its interaction with VirB10, and the incorporation of VirB2 and VirB5 into T-pili.
Keywords: Agrobacterium tumefaciens; T-pilus; bacteria; bacterial pathogenesis; membrane protein; protein secretion; protein–protein interaction; type IV secretion system; virulence; virulence factor.
© 2018 Mary et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

