Proteolytic ectodomain shedding of membrane proteins in mammals-hardware, concepts, and recent developments
- PMID: 29976761
- PMCID: PMC6068445
- DOI: 10.15252/embj.201899456
Proteolytic ectodomain shedding of membrane proteins in mammals-hardware, concepts, and recent developments
Abstract
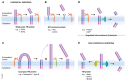
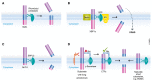
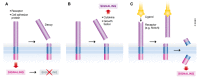
Proteolytic removal of membrane protein ectodomains (ectodomain shedding) is a post-translational modification that controls levels and function of hundreds of membrane proteins. The contributing proteases, referred to as sheddases, act as important molecular switches in processes ranging from signaling to cell adhesion. When deregulated, ectodomain shedding is linked to pathologies such as inflammation and Alzheimer's disease. While proteases of the "a disintegrin and metalloprotease" (ADAM) and "beta-site APP cleaving enzyme" (BACE) families are widely considered as sheddases, in recent years a much broader range of proteases, including intramembrane and soluble proteases, were shown to catalyze similar cleavage reactions. This review demonstrates that shedding is a fundamental process in cell biology and discusses the current understanding of sheddases and their substrates, molecular mechanisms and cellular localizations, as well as physiological functions of protein ectodomain shedding. Moreover, we provide an operational definition of shedding and highlight recent conceptual advances in the field. While new developments in proteomics facilitate substrate discovery, we expect that shedding is not a rare exception, but rather the rule for many membrane proteins, and that many more interesting shedding functions await discovery.
Keywords: matrix metalloproteases; meprin β; pro‐protein convertases; rhomboids; signal peptide peptidase‐like.
© 2018 The Authors.
Figures

Similar articles
-
Substrate requirements for SPPL2b-dependent regulated intramembrane proteolysis.J Biol Chem. 2009 Feb 27;284(9):5662-70. doi: 10.1074/jbc.M807485200. Epub 2008 Dec 29. J Biol Chem. 2009. PMID: 19114711
-
Meprin β induces activities of A disintegrin and metalloproteinases 9, 10, and 17 by specific prodomain cleavage.FASEB J. 2019 Nov;33(11):11925-11940. doi: 10.1096/fj.201801371R. Epub 2019 Aug 9. FASEB J. 2019. PMID: 31381863 Free PMC article.
-
The Coxsackievirus and Adenovirus Receptor (CAR) undergoes ectodomain shedding and regulated intramembrane proteolysis (RIP).PLoS One. 2013 Aug 28;8(8):e73296. doi: 10.1371/journal.pone.0073296. eCollection 2013. PLoS One. 2013. PMID: 24015300 Free PMC article.
-
Alpha-secretase activity of the disintegrin metalloprotease ADAM 10. Influences of domain structure.Ann N Y Acad Sci. 2000;920:215-22. doi: 10.1111/j.1749-6632.2000.tb06925.x. Ann N Y Acad Sci. 2000. PMID: 11193153 Review.
-
Physiological functions of SPP/SPPL intramembrane proteases.Cell Mol Life Sci. 2020 Aug;77(15):2959-2979. doi: 10.1007/s00018-020-03470-6. Epub 2020 Feb 12. Cell Mol Life Sci. 2020. PMID: 32052089 Free PMC article. Review.
Cited by
-
Mining oomycete proteomes for metalloproteases leads to identification of candidate virulence factors in Phytophthora infestans.Mol Plant Pathol. 2021 May;22(5):551-563. doi: 10.1111/mpp.13043. Epub 2021 Mar 3. Mol Plant Pathol. 2021. PMID: 33657266 Free PMC article.
-
Members of the Fibroblast Growth Factor Receptor Superfamily Are Proteolytically Cleaved by Two Differently Activated Metalloproteases.Int J Mol Sci. 2021 Mar 20;22(6):3165. doi: 10.3390/ijms22063165. Int J Mol Sci. 2021. PMID: 33804608 Free PMC article.
-
LRRK2 regulates endoplasmic reticulum-mitochondrial tethering through the PERK-mediated ubiquitination pathway.EMBO J. 2020 Jan 15;39(2):e100875. doi: 10.15252/embj.2018100875. Epub 2019 Dec 10. EMBO J. 2020. PMID: 31821596 Free PMC article.
-
Proteomic Analysis of MYB-Regulated Secretome Identifies Functional Pathways and Biomarkers: Potential Pathobiological and Clinical Implications.J Proteome Res. 2020 Feb 7;19(2):794-804. doi: 10.1021/acs.jproteome.9b00641. Epub 2020 Jan 27. J Proteome Res. 2020. PMID: 31928012 Free PMC article.
-
NrCAM is a marker for substrate-selective activation of ADAM10 in Alzheimer's disease.EMBO Mol Med. 2019 Apr;11(4):e9695. doi: 10.15252/emmm.201809695. EMBO Mol Med. 2019. PMID: 30833305 Free PMC article.
References
-
- Althoff K, Reddy P, Voltz N, Rose‐John S, Mullberg J (2000) Shedding of interleukin‐6 receptor and tumor necrosis factor alpha. Contribution of the stalk sequence to the cleavage pattern of transmembrane proteins. Eur J Biochem 267: 2624–2631 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

