Principles for targeting RNA with drug-like small molecules
- PMID: 29977051
- PMCID: PMC6420209
- DOI: 10.1038/nrd.2018.93
Principles for targeting RNA with drug-like small molecules
Abstract
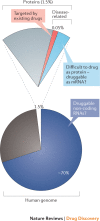
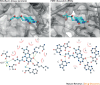
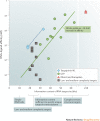
RNA molecules are essential for cellular information transfer and gene regulation, and RNAs have been implicated in many human diseases. Messenger and non-coding RNAs contain highly structured elements, and evidence suggests that many of these structures are important for function. Targeting these RNAs with small molecules offers opportunities to therapeutically modulate numerous cellular processes, including those linked to 'undruggable' protein targets. Despite this promise, there is currently only a single class of human-designed small molecules that target RNA used clinically - the linezolid antibiotics. However, a growing number of small-molecule RNA ligands are being identified, leading to burgeoning interest in the field. Here, we discuss principles for discovering small-molecule drugs that target RNA and argue that the overarching challenge is to identify appropriate target structures - namely, in disease-causing RNAs that have high information content and, consequently, appropriate ligand-binding pockets. If focus is placed on such druggable binding sites in RNA, extensive knowledge of the typical physicochemical properties of drug-like small molecules could then enable small-molecule drug discovery for RNA targets to become (only) roughly as difficult as for protein targets.
Conflict of interest statement
K.M.W. is an adviser to and holds equity in Ribometrix. K.D.W. and C.E.H. are employees of Ribometrix.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

