PCR-based detection of Plasmodium falciparum in saliva using mitochondrial cox3 and varATS primers
- PMID: 29977122
- PMCID: PMC6013985
- DOI: 10.1186/s41182-018-0100-2
PCR-based detection of Plasmodium falciparum in saliva using mitochondrial cox3 and varATS primers
Abstract
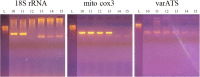
Background: Sampling of saliva for diagnosing Plasmodium falciparum infections is a safe, non-invasive alternative to sampling of blood. However, the use of saliva presents a challenge because lower concentrations of parasite DNA are present in saliva compared to peripheral blood. Therefore, a sensitive method is needed for detection of parasite DNA in saliva. This study utilized two recently reported "ultra-sensitive" PCR assays based on detection of the P. falciparum mitochondrial cox3 gene and the multi-copy nuclear varATS gene. The ultra-sensitive assays have an advantage over standard 18S rRNA gene-based PCR assay as they target genes with higher copy numbers per parasite genome. Stored saliva DNA samples from 60 Cameroonian individuals with infections previously confirmed by 18S rRNA gene PCR in peripheral blood were tested with assays targeting the cox3 and varATS genes.
Results: Overall, the standard 18S rRNA gene-based PCR assay detected P. falciparum DNA in 62% of the stored saliva DNA samples, whereas 77 and 68% of the samples were positive with assays that target the cox3 and varATS genes, respectively. Interestingly, the ultra-sensitive assays detected more P. falciparum infections in stored saliva samples than were originally detected by thick-film microscopy (41/60 = 68%). When stratified by number of parasites in the blood, the cox3 assay successfully detected more than 90% of infections using saliva when individuals had > 1000 parasites/μl of peripheral blood, but sensitivity was reduced at submicroscopic parasitemia levels. Bands on electrophoresis gels were distinct for the cox3 assay, whereas faint or non-specific bands were sometimes observed for varATS and 18S rRNA that made interpretation of results difficult. Assays could be completed in 3.5 and 3 h for the cox3 and varATS assays, respectively, whereas the 18S rRNA gene assays required at least 7 h.
Conclusions: This study demonstrates that a PCR assay targeting the cox3 gene detected P. falciparum DNA in more saliva samples than primers for the 18S rRNA gene. Non-invasive collection of saliva in combination with the proposed cox3 primer-based PCR assay could potentially enhance routine testing of P. falciparum during disease surveillance, monitoring, and evaluation of interventions for malaria elimination.
Keywords: 18S rRNA; Cameroon; Detection; Malaria diagnosis; Malaria surveillance; Mitochondrial genome; Plasmodium falciparum; Saliva-based assays; cox3; varATS.
Conflict of interest statement
Blood and saliva samples used were deidentified, archival samples collected in 2015. The current study was exempt from human subject research by the Committee on Human Studies, University of Hawaii at Manoa (Protocol Number: 2017-00395).The authors declare that they have no competing interests.Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Ultra-sensitive detection of Plasmodium falciparum by amplification of multi-copy subtelomeric targets.PLoS Med. 2015 Mar 3;12(3):e1001788. doi: 10.1371/journal.pmed.1001788. eCollection 2015 Mar. PLoS Med. 2015. PMID: 25734259 Free PMC article.
-
Dry season prevalence of Plasmodium falciparum in asymptomatic gambian children, with a comparative evaluation of diagnostic methods.Malar J. 2022 Jun 7;21(1):171. doi: 10.1186/s12936-022-04184-9. Malar J. 2022. PMID: 35672850 Free PMC article.
-
Comparison of three PCR-based assays for the non-invasive diagnosis of malaria: detection of Plasmodium parasites in blood and saliva.Eur J Clin Microbiol Infect Dis. 2014 Sep;33(9):1631-9. doi: 10.1007/s10096-014-2121-z. Epub 2014 May 5. Eur J Clin Microbiol Infect Dis. 2014. PMID: 24792127
-
Detection of Plasmodium falciparum DNA in saliva samples stored at room temperature: potential for a non-invasive saliva-based diagnostic test for malaria.Malar J. 2017 Oct 27;16(1):434. doi: 10.1186/s12936-017-2084-5. Malar J. 2017. PMID: 29078786 Free PMC article.
-
Utility of ultra-sensitive qPCR to detect Plasmodium falciparum and Plasmodium vivax infections under different transmission intensities.Malar J. 2020 Sep 3;19(1):319. doi: 10.1186/s12936-020-03374-7. Malar J. 2020. PMID: 32883308 Free PMC article.
Cited by
-
Does Antibody Avidity to Plasmodium falciparum Merozoite Antigens Increase with Age in Individuals Living in Malaria-Endemic Areas?Infect Immun. 2021 May 17;89(6):e00522-20. doi: 10.1128/IAI.00522-20. Print 2021 May 17. Infect Immun. 2021. PMID: 33722929 Free PMC article.
-
Zoonotic Blood-Borne Pathogens in Non-Human Primates in the Neotropical Region: A Systematic Review.Pathogens. 2021 Aug 10;10(8):1009. doi: 10.3390/pathogens10081009. Pathogens. 2021. PMID: 34451473 Free PMC article. Review.
-
Development of new real-time PCR assays for detection and species differentiation of Plasmodium ovale.PLoS Negl Trop Dis. 2024 Sep 10;18(9):e0011759. doi: 10.1371/journal.pntd.0011759. eCollection 2024 Sep. PLoS Negl Trop Dis. 2024. PMID: 39255325 Free PMC article.
-
Saliva-based methods for SARS-CoV-2 testing in low- and middle-income countries.Bull World Health Organ. 2022 Dec 1;100(12):808-814. doi: 10.2471/BLT.22.288526. Epub 2022 Oct 3. Bull World Health Organ. 2022. PMID: 36466209 Free PMC article.
-
Improving the Molecular Diagnosis of Malaria: Droplet Digital PCR-Based Method Using Saliva as a DNA Source.Front Microbiol. 2022 May 13;13:882530. doi: 10.3389/fmicb.2022.882530. eCollection 2022. Front Microbiol. 2022. PMID: 35633683 Free PMC article.
References
-
- Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH. A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT) Am J Trop Med Hyg. 2007;77(6 Suppl):119–127. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
