Activation of the endoplasmic reticulum stress sensor IRE1α by the vaccine adjuvant AS03 contributes to its immunostimulatory properties
- PMID: 29977610
- PMCID: PMC6023910
- DOI: 10.1038/s41541-018-0058-4
Activation of the endoplasmic reticulum stress sensor IRE1α by the vaccine adjuvant AS03 contributes to its immunostimulatory properties
Abstract
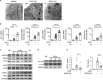
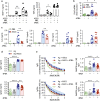
The oil-in-water emulsion Adjuvant System 03 (AS03) is one of the few adjuvants used in licensed vaccines. Previous work indicates that AS03 induces a local and transient inflammatory response that contributes to its adjuvant effect. However, the molecular mechanisms involved in its immunostimulatory properties are ill-defined. Upon intramuscular injection in mice, AS03 elicited a rapid and transient downregulation of lipid metabolism-related genes in the draining lymph node. In vitro, these modifications were associated with profound changes in lipid composition, alteration of endoplasmic reticulum (ER) morphology and activation of the unfolded protein response pathway. In vivo, treatment with a chemical chaperone or deletion of the ER stress sensor kinase IRE1α in myeloid cells decreased AS03-induced cytokine production and its capacity to elicit high affinity antigen-specific antibodies. In summary, our results indicate that IRE1α is a sensor for the metabolic changes induced by AS03 in monocytic cells and may constitute a canonical pathway that could be exploited for the design of novel vaccine adjuvants.
Conflict of interest statement
All authors have declared the following interests: R.G., C.C. and A.D. are employees of the GSK group of companies. A.M.D. reports ownership of GSK shares. The remaining authors declare no competing interests.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

